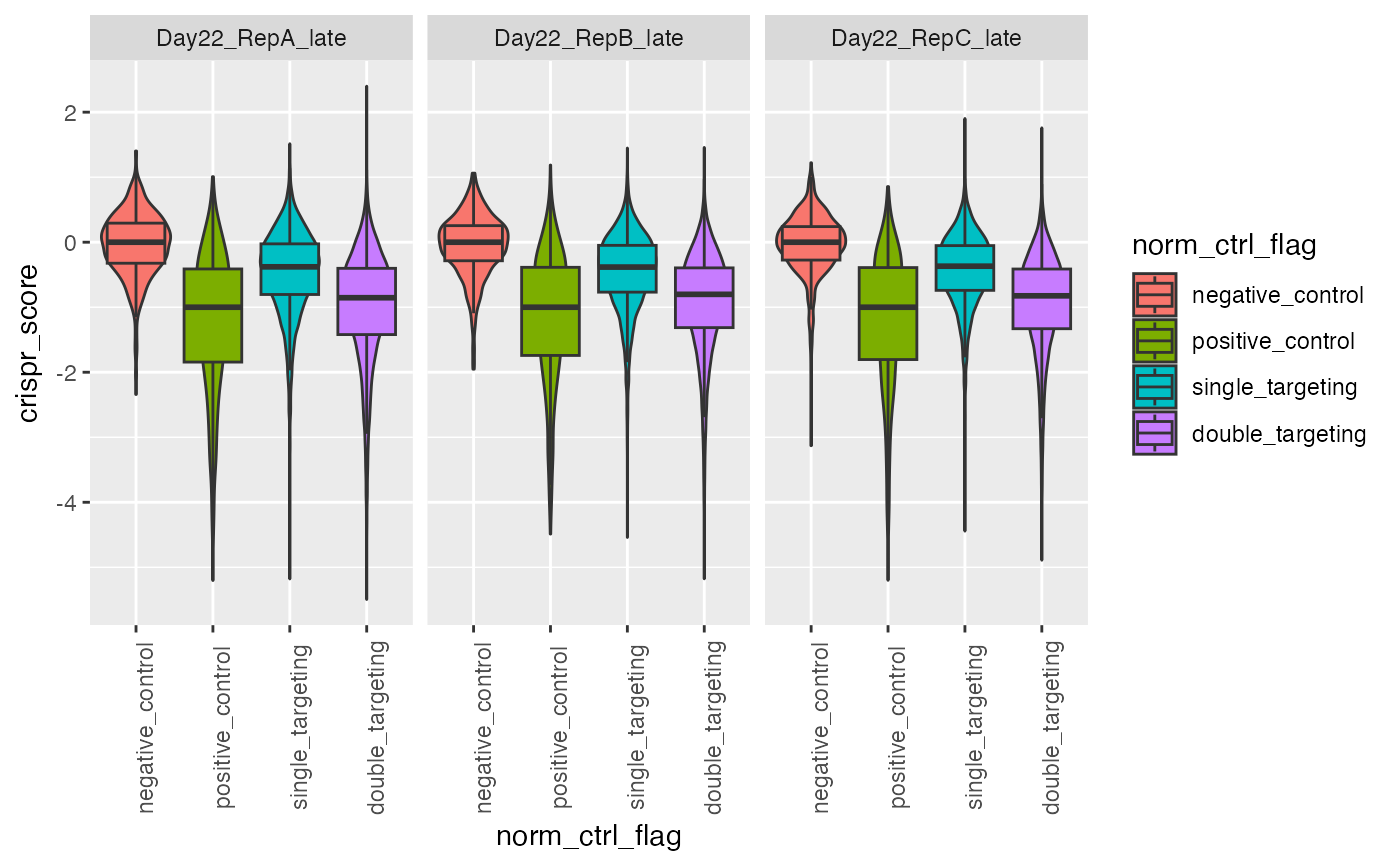
This plots normalization after CRISPR scores have been calculated
plot_crispr(.data = NULL, gimap_dataset, output_file = "crispr_norm_plot.png")Arguments
Value
A ggplot2 boxplot of the CRISPR scores separated by the type of target. Can be used to determine the normalization has proceeded properly.
Examples
# \donttest{
gimap_dataset <- get_example_data("gimap") %>%
gimap_filter() %>%
gimap_annotate(cell_line = "HELA") %>%
gimap_normalize(
timepoints = "day",
missing_ids_file = tempfile()
)
#> Annotating Data
#>
Downloading: 3.4 kB
Downloading: 3.4 kB
Downloading: 3.4 kB
Downloading: 3.4 kB
#> Downloading: Achilles_common_essentials.csv
#>
|
| | 0%
|
|======================================================================| 100%
#>
|
| | 0%
|
|=============================================== | 67%
|
|======================================================================| 100%
#> Normalizing Log Fold Change
# Plot:
plot_crispr(gimap_dataset)
 # }
# }