7 5’ UTR Free Energy Evaluation
To obtain the 5’ UTR sequences and their corresponding minimal free energy (MEF) values, we utilized the ShortRead, BioStrings, and BSgenome.Hsapiens.UCSC.hg38 Bioconductor packages, along with the Vienna software. The 5’ UTR models were based on transcripts, but since there could be multiple isoforms with the same 5’ UTR model, we ensured uniqueness by retaining only one instance of each.
In this study, we calculated the free energy for five sets of 5’ UTR features, as follows:
All annotated 5’ UTR: This set included transcript-based and unique 5’ UTR models extracted from Gencode v3.5, comprising a total of 19,570 genes.
DUX4 direct targets: We identified 84 DUX4 directed targets from Polysome-seq data, many of which had alternative promoters identified by Whiddon 2017. These targets were listed in
../extdata/DUX4-targets-novel-and-annotated-5'UTR.xlsx. We also checked for alternative splice events, alternative first exon (AFE), or alternative start site (AS) using../scripts/110-HDUX4-promoter.R.TE down Ribo-seq: Down-regulated translational efficiency by Ribo-seq (S6) in 5’UTR region: We identified 1,131 candidates with reduced translational efficiency in the 5’UTR region based on Ribo-seq data.
Poly-seq up-regulated: We identified 239 non-DUX4-induced candidates that were up-regulated in DUX4-treatment in Poly-seq samples (High/High).
Poly-seq down-regulated: We identified 5,800 candidates that were down-regulated in Poly-seq samples in the High vs. Sub comparison.
The plan of calculating the the five collections of 5’UTR is implemented by 09B-5UTR-structure.R.
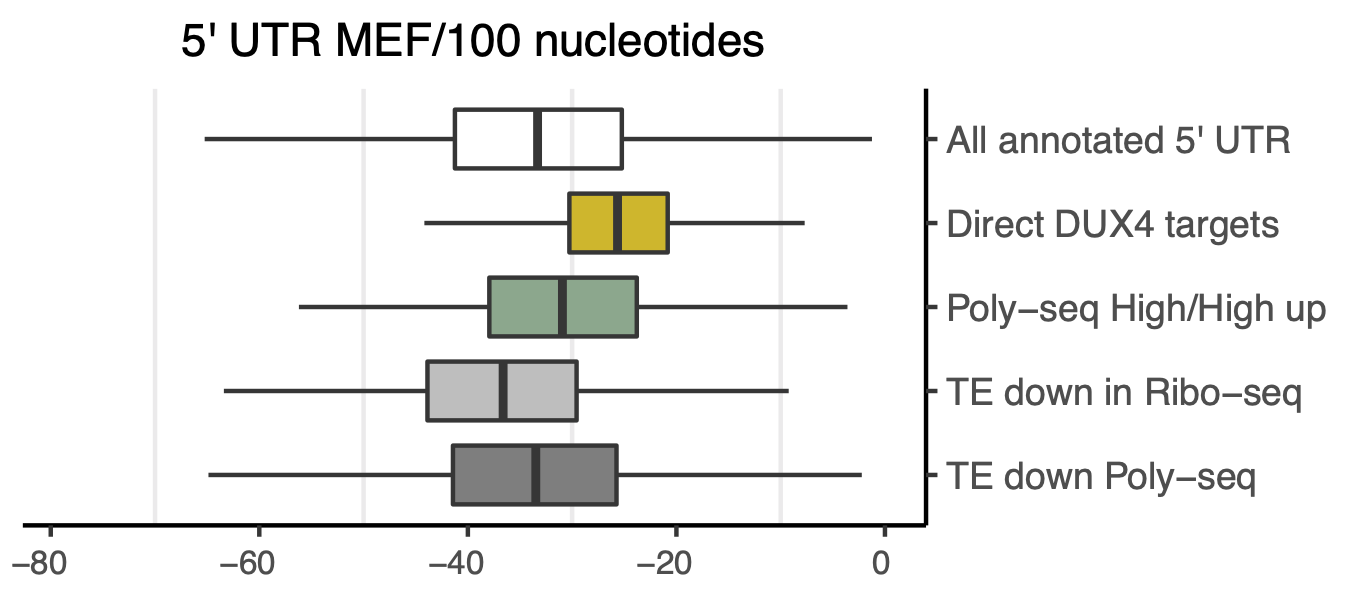
Figure 7.1: 5’ UTR minimal free energy per 100 nucleotides