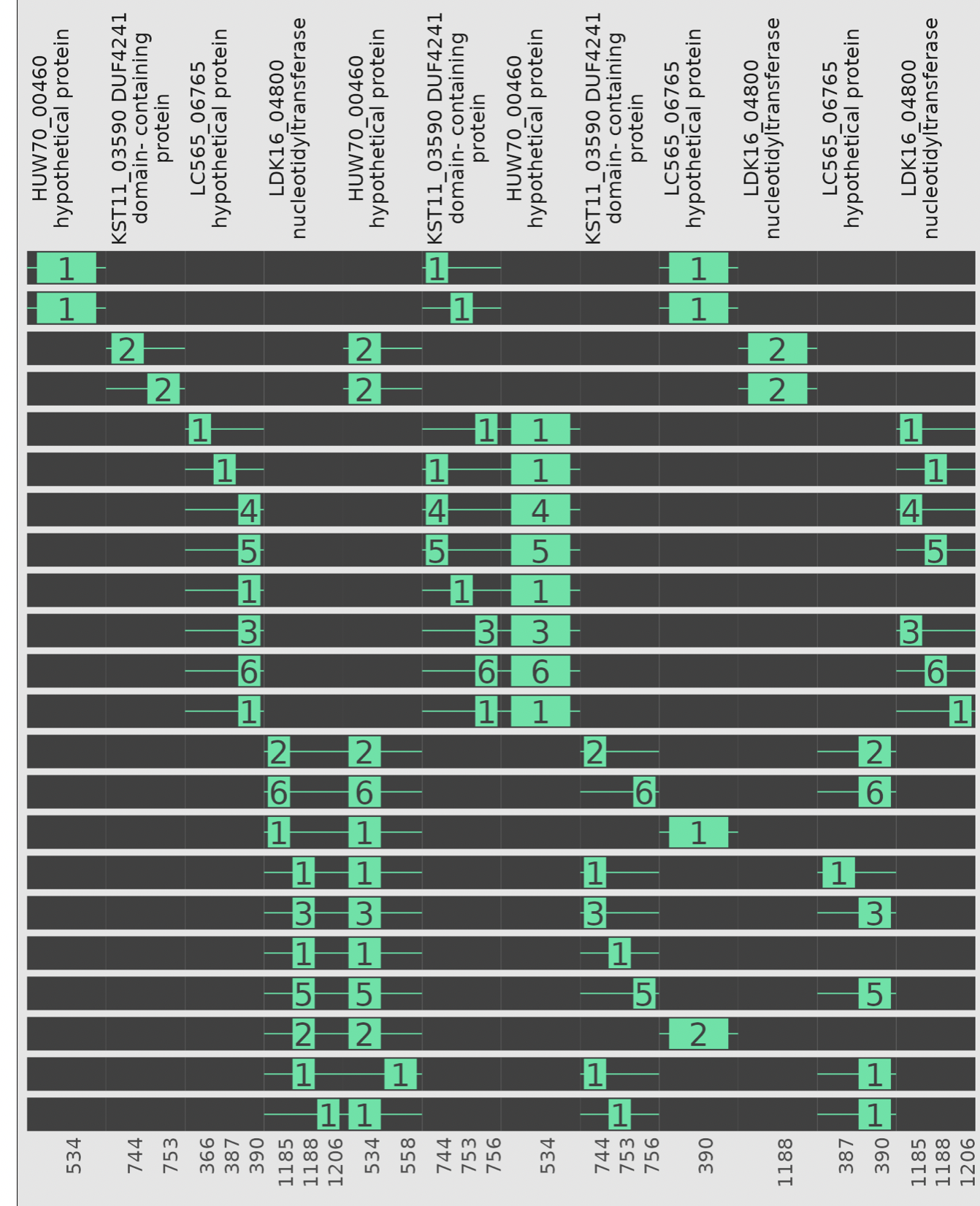
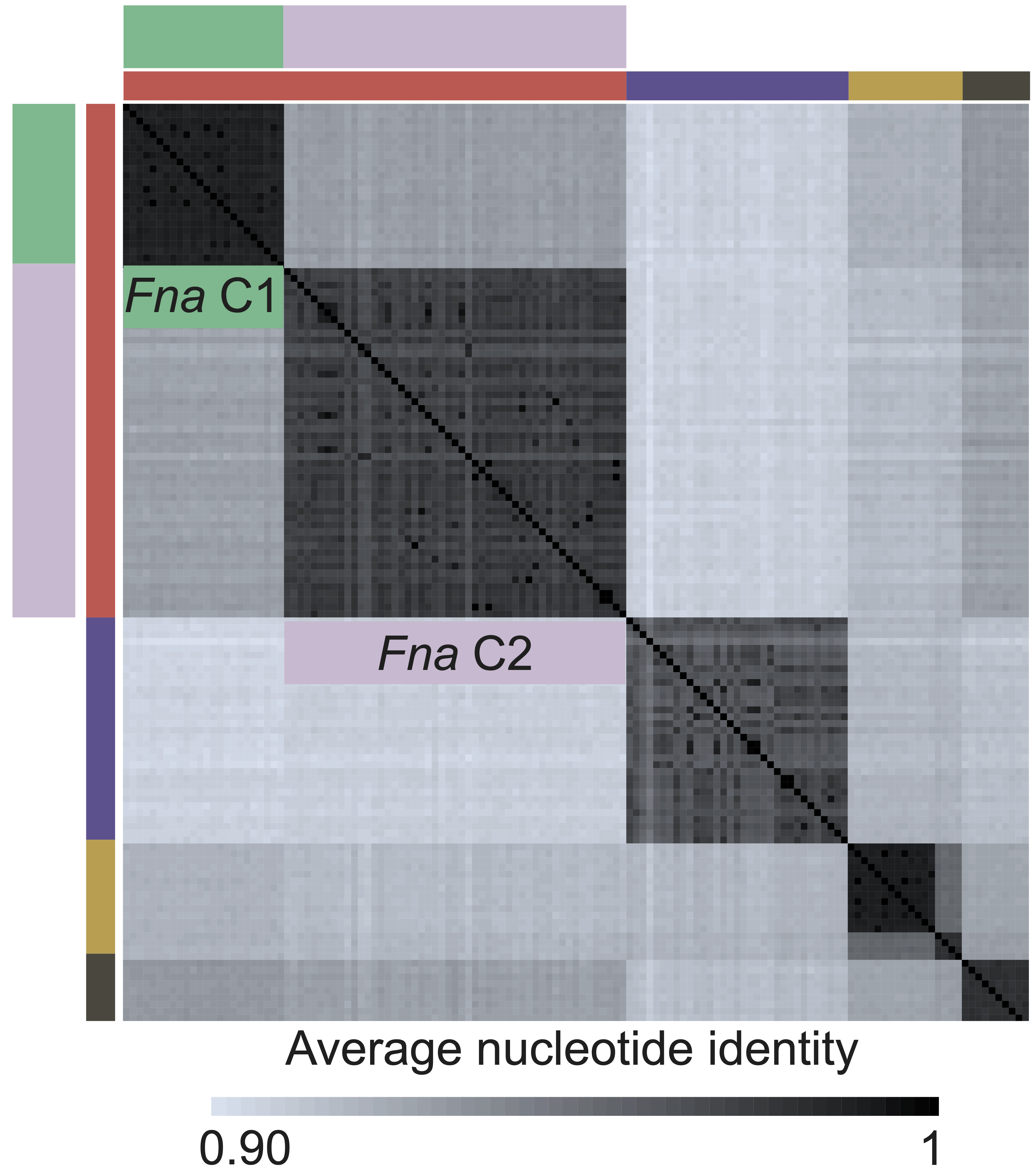
GiG Map
A heatmap resulting from the alignment of a set of genes against a set of genomes (to create a similarity score) alongside a phylogenetic tree of genome similarity. Additional information is shown by adding categorical annotations of the genomes.
To learn more or create your own GiG Map.