Chapter 5 DUXC transcriptome compared to early embryo transcriptome profile
We compared DUXC transcriptome to homologs of the early mouse and human embryo transcriptome to determine whether DUXC transcritome is enriched for genes expressed in the early embryo. Using the homologs of mouse 2C-like signature (Tomohiko Akiyama and Ko 2015), GSEA suggested that that DUXC transcriptome is enriched for genes expressed in the homologs of mouse 2C-like.
Load datasets
pkg_dir <- "~/CompBio/canFam3.DuxFamily"
source(file.path(pkg_dir, "scripts", "load_lib_dataset.R"))
suppressPackageStartupMessages(library(latex2exp))
load(file.path(pkg_dir, "data", "canine_human_mouse_ensOrtholog.rda"))
load(file.path(pkg_dir, "data", "human_mouse_ensOrtholog.rda"))Tool: hypergeometric test
# hypergeometric test
.hypergeometric <- function(x, k, universe, selected) {
white_ball <- length(selected)
black_ball <- length(universe) - white_ball
prob <- dhyper(x=x, m=white_ball, n=black_ball, k=k, log = FALSE)
expected <- k * white_ball/(white_ball+black_ball)
return(c(total=black_ball+white_ball, up_regulated=white_ball,
gene_set=k, up_reg_geneset=x,
expected=expected, prob=prob))
}5.1 Mouse 2C-like signature
Out of 466 genes from mouse 2C-like signature, 292 have homologs in canine. 58 out 292 (20%) are positively affected by DUXC in canine; GSEA gave p-value \(7e-9\) suggested that DUXC transcriptome is enriched for the genes of 2C-like signature.
# get 2C-like gene signature (Akiyama 2015) and homologs in canine
load(file.path(data_dir, "z4_ensembl.rda"))
z4_ensembl <- z4_ensembl %>% rename(MOUSE_ENSEMBL = ENSEMBL)
mouse2C_homolog <- canine_human_mouse_ensOrtholog %>%
#dplyr::select(ensembl_gene_id, mmusculus_homolog_ensembl_gene, mmusculus_homolog_associated_gene_name) %>%
rename(CANINE_ENSEMBL=ensembl_gene_id, MOUSE_ENSEMBL=mmusculus_homolog_ensembl_gene,
HUMAN_ENSEMBL=hsapiens_homolog_ensembl_gene) %>%
dplyr::filter(MOUSE_ENSEMBL != "", !duplicated(MOUSE_ENSEMBL), !duplicated(CANINE_ENSEMBL)) %>%
inner_join(z4_ensembl, by="MOUSE_ENSEMBL") %>%
left_join(CinC, by="CANINE_ENSEMBL") %>%
left_join(MinM, by="MOUSE_ENSEMBL") %>%
left_join(HinH, by="HUMAN_ENSEMBL") %>%
dplyr::mutate(CinC_up = CinC_padj < 0.05 & CinC_logFC > 0) %>%
dplyr::mutate(MinM_up = MinM_padj < 0.05 & MinM_logFC > 0) %>%
dplyr::mutate(HinH_up = HinH_padj < 0.05 & HinH_logFC > 0)
universe <- pull(CinC, CANINE_ENSEMBL)
selected <- CinC %>% dplyr::filter(CinC_logFC > 0, CinC_padj < 0.05) %>% pull(CANINE_ENSEMBL)
x <- sum(mouse2C_homolog$CinC_up, na.rm=TRUE)
k <- nrow(mouse2C_homolog)
prob <- .hypergeometric(x=x, k=k, universe=universe, selected=selected)
prob## total up_regulated gene_set up_reg_geneset expected
## 1.70750e+04 1.56200e+03 2.92000e+02 5.80000e+01 2.67118e+01
## prob
## 6.99034e-09pearson <- mouse2C_homolog %>%
dplyr::filter(!is.na(MinM_logFC), !is.na(CinC_logFC)) %>%
summarise(pearson = cor(MinM_logFC, CinC_logFC))
label <- sprintf("Up-regulated in CinC = %2.0f / %1.0f \npearson = %0.2f \nGSEA p-value < 7e-9",
sum(mouse2C_homolog$CinC_up, na.rm=TRUE),
nrow(mouse2C_homolog), pearson)
ggplot(mouse2C_homolog, aes(x=CinC_logFC, y=MinM_logFC, color=CinC_up)) +
#geom_abline(intercept=0, slope=1, color="gray75", alpha=0.2) +
geom_smooth(method = "lm", se=FALSE, color="gray75", linetype="dashed") +
geom_point(alpha=0.7, size=1.2) +
geom_text(x=-4.5, y=7.5, label=label, hjust=0, vjust=1,
aes(fontface="plain"), color="gray25") +
theme_bw() +
theme(legend.position="none") +
scale_color_manual(values=c("ivory4", "firebrick4")) +
labs(title="FSHD signature homologs: MinM vs. HinH",
y=TeX("MinM: $\\log_2$(DUX4/no DUX4) in human"),
x=TeX("CinC: $\\log_2$(Dux/no Dux) in mouse")) +
theme(plot.title = element_text(hjust = 0.5, face="bold"))## `geom_smooth()` using formula 'y ~ x'## Warning: Removed 114 rows containing non-finite values (stat_smooth).## Warning: Removed 114 rows containing missing values (geom_point).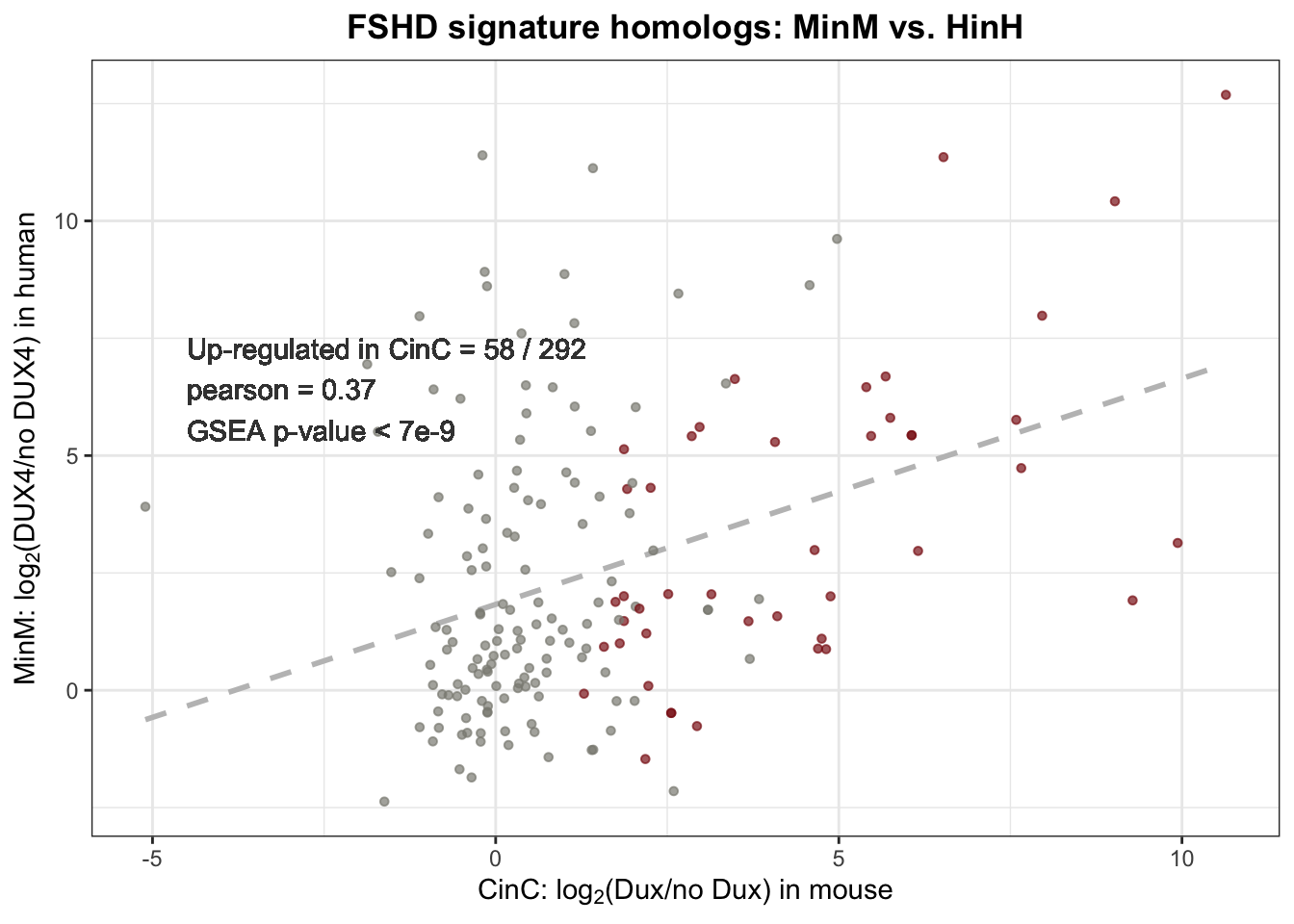
ggsave(filename=file.path(pkg_dir, "figures",
"MinM_CinC_2Clike_signature_homologs.pdf"),
width=5, height=3.75)## `geom_smooth()` using formula 'y ~ x'## Warning: Removed 114 rows containing non-finite values (stat_smooth).
## Warning: Removed 114 rows containing missing values (geom_point).5.2 cleavage-stage signature
We also tested if DUXC transcriptome is enriched for genes expressed from human cleavage-stage signature (Hendrickson 2017). Out of 738 genes from human cleavage-stage signature, 380 has homologs in canine. Only 44 out of the 380 homologs are up-regulated by DUXC in canine (12%); the \(p\)-value from GSEA is 0.18, not significant to suggest an enrichment for homologs of human cleavage-stage signature.
# luster 4 identified in Hendrickson 2017: cleavage-stage signature
load(file.path(data_dir, "human_cleavage.rda"))
human_cleavage <- human_cleavage %>% rename(HUMAN_ENSEMBL = GeneID)
# get human-canine cleavage homologs: 380
cleavage_homolog <- canine_human_mouse_ensOrtholog %>%
rename(CANINE_ENSEMBL=ensembl_gene_id, HUMAN_ENSEMBL=hsapiens_homolog_ensembl_gene,
MOUSE_ENSEMBL=mmusculus_homolog_ensembl_gene) %>%
dplyr::filter(HUMAN_ENSEMBL != "", !duplicated(HUMAN_ENSEMBL), !duplicated(CANINE_ENSEMBL)) %>%
inner_join(human_cleavage, by="HUMAN_ENSEMBL") %>%
left_join(CinC, by="CANINE_ENSEMBL") %>%
left_join(HinH, by="HUMAN_ENSEMBL") %>%
left_join(MinM, by="MOUSE_ENSEMBL") %>%
dplyr::mutate(CinC_up = CinC_padj < 0.05 & CinC_logFC > 0) %>%
dplyr::mutate(HinH_up = HinH_padj < 0.05 & HinH_logFC > 0) %>%
dplyr::mutate(MinM_up = MinM_padj < 0.05 & MinM_logFC > 0)
# CinC GSEA hypergeometric
universe <- pull(CinC, CANINE_ENSEMBL)
selected <- CinC %>% dplyr::filter(CinC_logFC > 0, CinC_padj < 0.05) %>% pull(CANINE_ENSEMBL)
x <- sum(cleavage_homolog$CinC_up, na.rm=TRUE)
k <- nrow(cleavage_homolog)
prob <- .hypergeometric(x=x, k=k, universe=universe, selected=selected)
prob## total up_regulated gene_set up_reg_geneset expected
## 1.707500e+04 1.562000e+03 3.800000e+02 4.400000e+01 3.476193e+01
## prob
## 1.787702e-025.3 iPSCs
We used the DUX4 expressed iPSCs (???) as an early embryo model. The hygergeometric test yielded \(p-value < 2e-13\).
ipscs <- read_csv(file.path(pkg_dir, "extdata", "handrickson_2017_supple_table6.csv"),
skip_empty_rows=TRUE) %>%
dplyr::filter(`Lg2Rto DUX4_24hr_Luciferase_24hr` > 1, `AdjP DUX4_24hr_Luciferase_24hr` < 0.01) %>%
dplyr::select(ID, Gene, `Lg2Rto DUX4_24hr_Luciferase_24hr`, `AdjP DUX4_24hr_Luciferase_24hr` ) %>%
rename(HUMAN_ENSEMBL=ID, HinIPSCs_logFC = `Lg2Rto DUX4_24hr_Luciferase_24hr`,
HinIPSCs_padj=`AdjP DUX4_24hr_Luciferase_24hr`)
ipsc_homolog <- canine_human_mouse_ensOrtholog %>%
rename(CANINE_ENSEMBL=ensembl_gene_id, HUMAN_ENSEMBL=hsapiens_homolog_ensembl_gene,
MOUSE_ENSEMBL=mmusculus_homolog_ensembl_gene) %>%
dplyr::filter(HUMAN_ENSEMBL != "", !duplicated(HUMAN_ENSEMBL), !duplicated(CANINE_ENSEMBL)) %>%
inner_join(ipscs, by="HUMAN_ENSEMBL") %>%
left_join(CinC, by="CANINE_ENSEMBL") %>%
left_join(MinM, by="MOUSE_ENSEMBL") %>%
dplyr::mutate(CinC_up = CinC_padj < 0.05 & CinC_logFC > 0) %>%
dplyr::mutate(MinM_up = MinM_padj < 0.05 & MinM_logFC > 0)
# CinC GSEA hypergeometric
universe <- pull(CinC, CANINE_ENSEMBL)
selected <- CinC %>% dplyr::filter(CinC_logFC > 0, CinC_padj < 0.05) %>% pull(CANINE_ENSEMBL)
x <- sum(ipsc_homolog$CinC_up, na.rm=TRUE)
k <- nrow(ipsc_homolog)
prob <- .hypergeometric(x=x, k=k, universe=universe, selected=selected)
prob## total up_regulated gene_set up_reg_geneset expected
## 1.707500e+04 1.562000e+03 1.020000e+02 3.600000e+01 9.330835e+00
## prob
## 2.684079e-13# References
Tomohiko Akiyama, Mayumi Oda1, Li Xin, and Minoru S. H. Ko. 2015. “Transient Bursts of Zscan4 Expression Are Accompanied by the Rapid Derepression of Heterochromatin in Mouse Embryonic Stem Cells.” DNA Research, no. 22.