Chapter 8 Non-LTR retrotrasposons and satellites
This chapter investigated the satellites and non-LTR retrotransposons transcriptional activites. The non-LTR retrotransposons include short interspersed nuclear elements (SINEs) and long interspersed nuclear elements (LINEs).
Summary: non-LTR tretrotrasponses and satellites activiation in canine
Studies ((Sean C Shadle 2019) and (Jennifer L. Whiddon 2017)) have shown that in human myoblast, DUX4 enriched HSATII and families of non-LTR RNA transcripts, such as Alu family of SINE and LINE1 (L1). Therefore, we were interested in examing the conservation of the non-LTR activation by DUX4 in our canine model. Our analysis showed that DUX4 up-regulated 53% of L1s (52/97) and 28.5% of DUX4 binding sites overlap with L1s. Given that L1s constitute 17% of the canine genome, DUX4, whether directly interacted with L1s or not, induced the L1 family’s enrichment in canine myoblasts. On the other hand, the enrichment of L1s by DUXC was rather unimpressive - only 16% of L1s (16/97) exhibited differential expression, and 15.6% DUXC peaks overlap with L1s (figure 8.1).
Unlike human SINEs derived from 7SL-RNA, canine SINEs share no homology with humans (Bentolila 1999). They evolved from tRNA genes (Bentolila 1999) and are categorized by RMSK into tRNA-Lys (72%, SINEC_#), MIR (27%), Deu (0.2%, AmnSINE1, AmnSINE2), and tRNA (0.2%, LFSINE_Vert, MamSINE1) families (supplemental figure). Six out of 11 tRNA-Lys elements (54%) are up-regulated in DUX4 expressed cell line; in fact, all tRNA-Lys have logFC > 1. That is, the tRNA-Lys family is enriched in HinC transcriptome. Though none of the tRNA-Lys was called up-regulated in DUXC cells, their expression is highly correlated with that of in DUX4 expressed cell line (figure 8.4).
Canine (canFam3) has 5 major satellites and satellite-like family: Bs, Carnivore Satellite (CarSat1, CarSat2), Satellite canis Familiaris (SAT1_CF, SAT2_CF, SAT3_CF, SAT4_CF, and SAT6_CF), (CATTC)n, (GAATG)n and SUBTEL_sa. Bs, CarSat1, and SAT1_CF were differentially up-regulated in DUX4 expressed cells, whereas in DUXC expressed cells, Bs element was called up-regulated, and CarSat1 was moderately affected (adjusted p-value = 0.14).
Load and datasets and libraries
# define directory
pkg_dir <- "~/CompBio/canFam3.DuxFamily"
fig_dir <- file.path(pkg_dir, "figures")
data_dir <- file.path(pkg_dir, "data")
# load library
suppressPackageStartupMessages(library(DESeq2))
suppressPackageStartupMessages(library(tidyverse))
suppressPackageStartupMessages(library(tidyr))
suppressPackageStartupMessages(library(plyranges))
suppressPackageStartupMessages(library(ggrepel))
suppressPackageStartupMessages(library(purrr))
suppressPackageStartupMessages(library(canFam3.rmsk))
load(file.path("~/CompBio/hg38/hg38.rmsk.rda"))
suppressPackageStartupMessages(library(wesanderson)) # modern palettes
# load datasets and local functions
source(file.path(pkg_dir, "scripts", "tools.R"))
load(file.path(pkg_dir, "data", "HinC_rmsk.rda")) # made in chapter 6
load(file.path(pkg_dir, "data", "CinC_rmsk.rda")) # made in chapter 6
load(file.path(pkg_dir, "data", "CALTinC_rmsk.rda")) # made in chapter 6
load(file.path(pkg_dir, "data", "HinH_rmsk.rda"))
load(file.path(data_dir, "peaks_list.rda"))Tidy up the differential analysis results on the repeats of HinC and CinC transcriptome into one data.frame instance.
comb <- inner_join(HinC_rmsk$res_df, CinC_rmsk$res_df,
by="repName", suffix=c("_HinC", "_CinC")) %>%
dplyr::mutate(DE_status = "neither") %>%
dplyr::mutate(DE_status =
replace(DE_status, padj_HinC < 0.05 | padj_CinC < 0.05, "either")) %>%
dplyr::mutate(DE_status = replace(DE_status, padj_HinC < 0.05 & padj_CinC < 0.05, "both")) %>%
dplyr::mutate(DE_status = factor(DE_status, levels=c("neither", "either", "both"))) reference: Analysis of major repetitive DNA sequences in the dog (Canis familiaris) genome, Betolila 1999.
8.1 LINEs
Background: LINEs constitute 20% of canine genome; LINE1 (L1) and LINE2 (L2) families derive 81% and 16% of the LINE repeat class, respectively. Whidden et al. (Jennifer L. Whiddon 2017) showed that DUX4 up-regulated, directly or indirectly, impressive numbers of LINE1s in human myoblast. In this project, we examed whether DUX4 and DUXC transcription activities lead to similar LINE1s activation in canine myblast cells.
Results:
- DUX4 up-regulated > 50% of L1s and L2s in canine cells
- 35% DUX4 peaks binds to LINEs (monoclonal cell line)
- GSEA suggested LINE1 family is enriched in DUX4 transcriptome in canine cells
- DUX4 up-regulated many carnivore-specific L1s and some Half-L1s (HAL1s, ancient L1-like)
- DUXC up-regulated some LINEs (16% in LINE1 family); GESA does not suggest L1 enrichment in DUXC canine cells
- Both DUX4 and DUXC up-regulate L1M2a (L1) and X6B_LINE (CR1) with impressive fold-change (~ 6 and 4)
- DUXC’s L1-overlapped peaks have a palindromic motif - AATCAATTGATT (243/691); DUX4’s have TAA(G/T/C)TCAATCA (431/999) (Chapter 9)
- Peaks overlapping with L1s have enriched AATCA sequence (Chapter 9)
## repFamily length prop ypos percent label
## 1 L1 412795245 81.219909588 40.60995 81% L1 (81%)
## 2 L2 82659880 16.263821014 89.35182 16% L2 (16%)
## 3 CR1 9758771 1.920095999 98.44378 2% CR1 ( 2%)
## 4 RTE-X 2869545 0.564599976 99.68613 1% RTE-X ( 1%)
## 5 Dong-R4 98128 0.019307265 99.97808 0% Dong-R4 ( 0%)
## 6 RTE-BovB 56831 0.011181836 99.99332 0% RTE-BovB ( 0%)
## 7 L1? 5511 0.001084322 99.99946 0% L1? ( 0%)DUX4 up-regulates > 50% of L1 and L2 retroelements in canine cells, whereas DUXC only up-regulated 16% of L1s. LFC of HinC and CinC are highly correlated.
## `summarise()` regrouping output by 'sample' (override with `.groups` argument)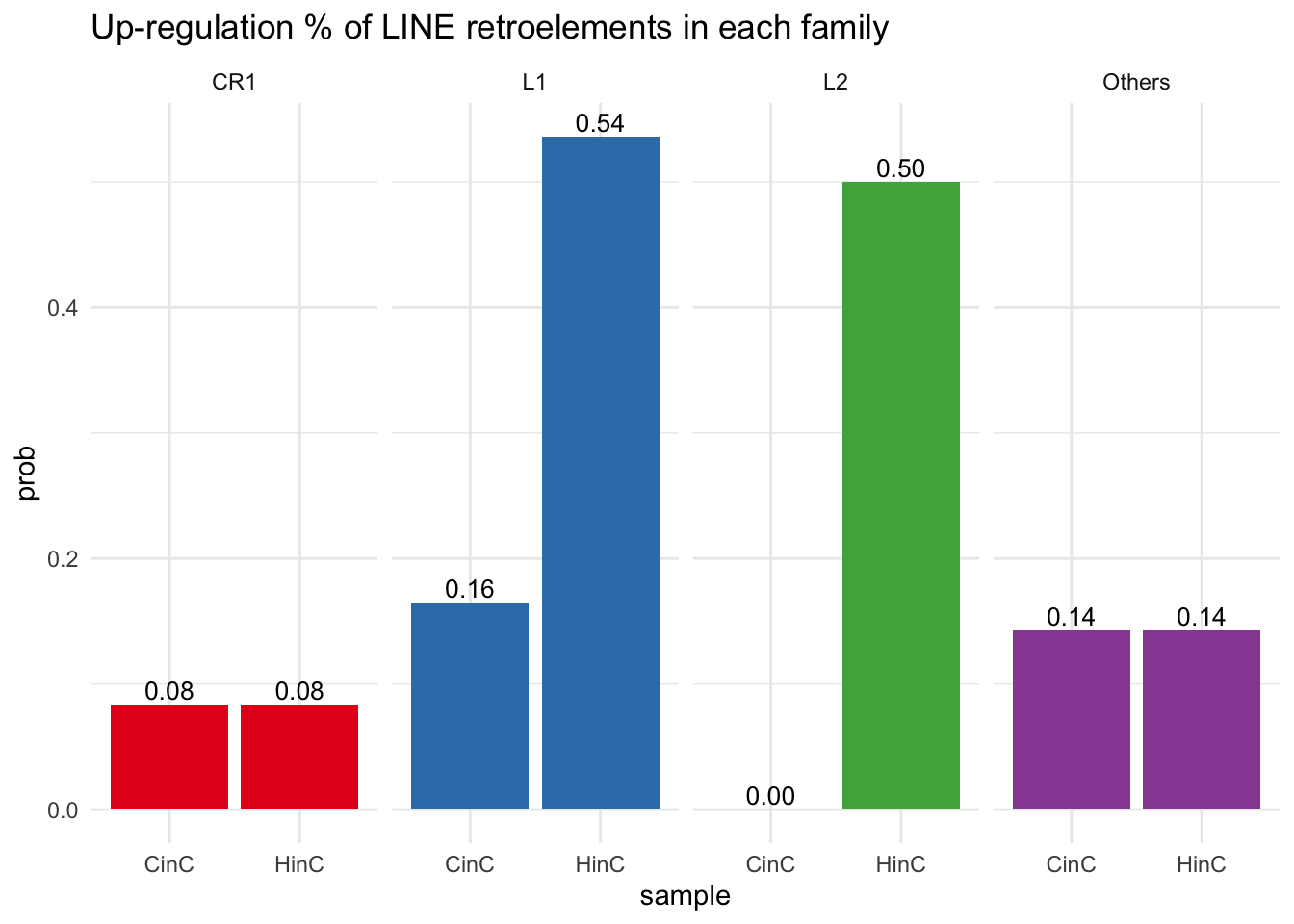
Figure 8.1: Percentage of up-regulation of LINE retroelements in each familiy. Others include unclassified L1, Dong-R4, RTE-BovB and RTR-X.
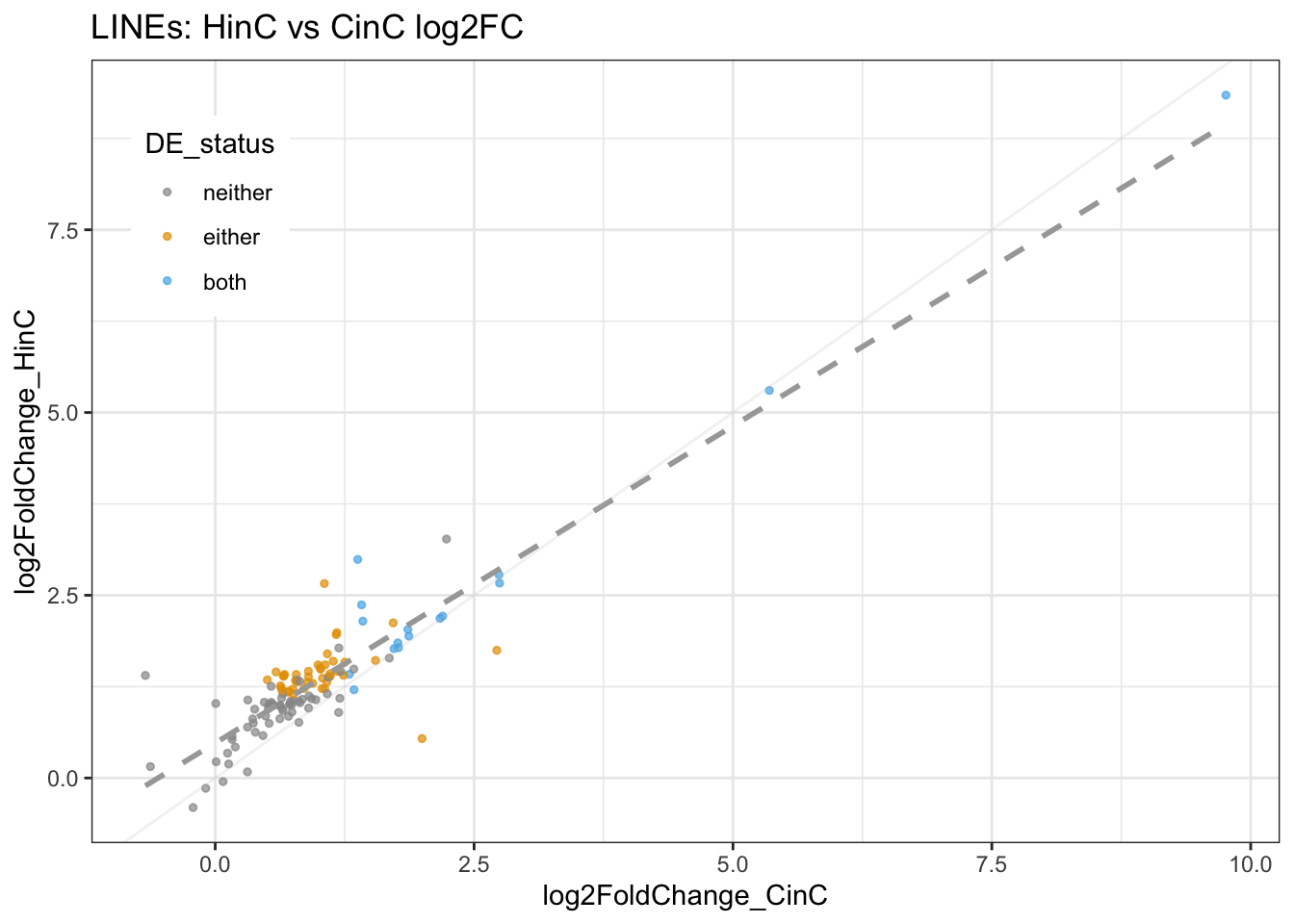
Figure 8.2: LINEs expression (LFC) in HinC and CinC.
8.1.1 L1s activation
Question: Which subfamilies of L1s were up-regulated by DUX4 in canine cells?
DUX4 up-regulated many mammalian-specific, carnivore-specific L1s and some HAL1s.
## Warning: Removed 10 rows containing missing values (position_stack).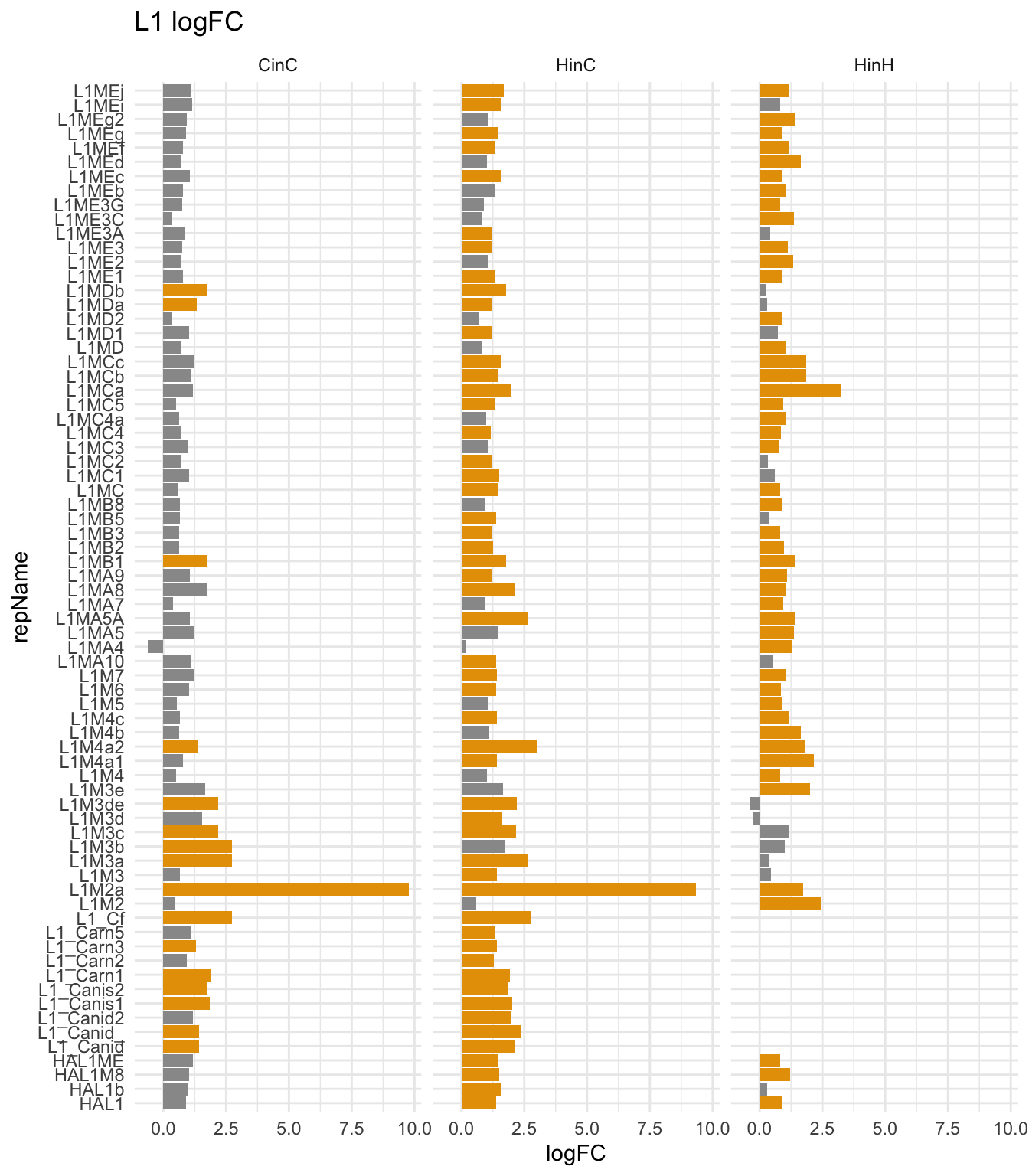
Figure 8.3: L1s activation in DUXC and DUX4 in canine cells. Only display significently expressio L1s in either CinC, HinC, or HinH.
8.1.2 LINE1 and DUXC/DUX4 binding sites
This section is a summary of Chapter 9.
Our previous study ((Jennifer L. Whiddon 2017)) and the canine model suggested that L1 subfamily is enriched in DUX4 expressed human and canine myoblasts. But does DUX4 (or DUXC) directly interact L1s? Are the binding sites on L1s artifacts? To answer the question, we want to first identify the motifs of peaks that overlap with the differentially expressed LINE1 elements (LFC > 1 with adjusted p-value < 0.05) by:
- use the narrowed peaks (+/- 50 bps from summit) and select the subset that overlap with the differentially expression L1s
- get sequence of the selected peaks
- use MEME to find the DUXC/DUX4 motif of these peaks
- use BioStrings::matchPattern() to find the motif enrichment
Results: 20.5% (21,888 out of 85,825) of DUX4 narrowed peaks overlap with L1s (25.50%); 14% of DUXC narrowed peaks overlap with L1s. Based on the top 1,000 of these L1-overlapped narrowed peaks of DUXC and DUX4 (on monoclonal cell lines), MEME identified a palindromic motif AATCAATTGATT (243/691) in DUXC cells and a motif TAAGTCAATCAG (431/999) in DUX4 cells. That is L1s do not have DUX4 nor DUXC motif enrichment; most likely that L1’s up-regulated are directly affected by DUX4 or DUXC in canine cells. Interestingly, however, both motifs of L1-overlapped peaks in DUX4 and DUXC cells obtain a motif AATCA, which is the canonical, second part of DUX4, Dux and DUXC motifs. This implies that L1s are enriched with AATCA sequences: 42.8% and 46.6% of DUXC and DUX4’s L1-overlapped narrowed peaks have AATCA sequences, mostly distributed near the center of the summits (technical details in Chapter 9).
Take away: DUXC and DUX4 seem not directly regulated L1s.
8.2 SINE
Background: The short interspersed repetitive elements (SINEs) constitute approximately 10% of canine genome. Canine SINEs evolved from tRNA genes whereas human SINEs evolved from 7SL-RNA, which gave rise to human Alu family (Simone Bentolila 1999); they do not share homology. However, known that human Alu family are enriched by DUX4 transcriptome in human myoblast (???), we are interested to know whether canine SINEs are also enriched by DUX4 and DUXC in canine myoblast and why?
Results: RMSK categorized canine SINEs into tRNA-Lys (72%, SINEC_*), MIR (27%), Deu (0.2%, AmnSINE1, AmnSINE2), and tRNA (0.2%, LFSINE_Vert, MamSINE1) families (Figure 8.4) with total 20 name/group and 1,610,013 instances of sequence. Six out of 11 tRNA-Lys (54%, named SINEC_*) are up-regulated in DUX4 expressed cell line, and all tRNA-Lys have logFC > 1. This means the tRNA-Lys family is enriched in HinC transcriptome. Though none of tRNA-Lys elements were called up-regulated by DUXC, their expression is highly correlated with that of in DUX4 expressed cell line.
Note: Murine SINEs are derived from both 7SL-RNA (B1 family) and tRNA (Simone Bentolila 1999).
8.2.1 Distribution of SINE family
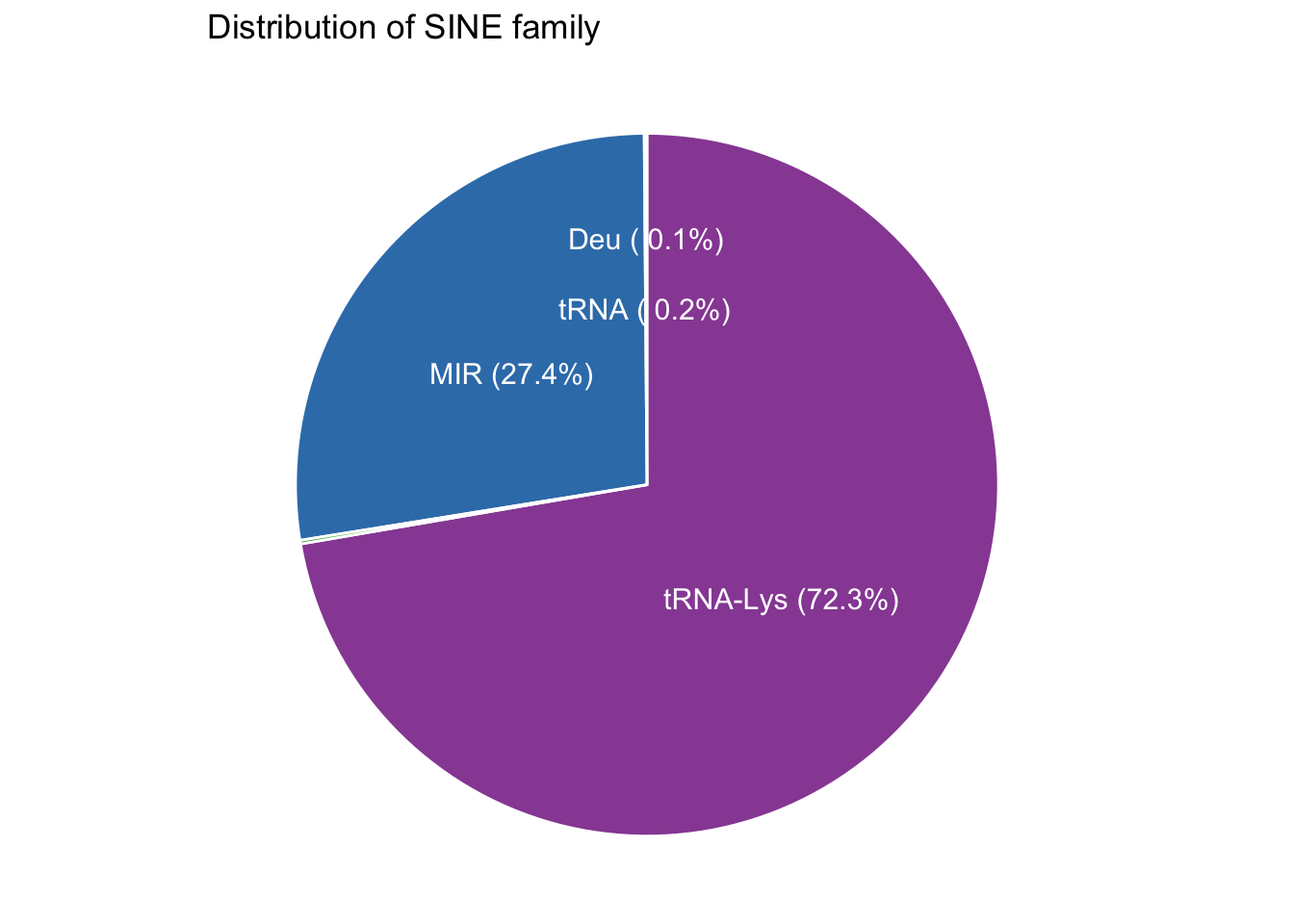
Figure 8.4: Distribution of SINE family.
8.2.2 Expression of SINEs in HinC and CinC transcriptome
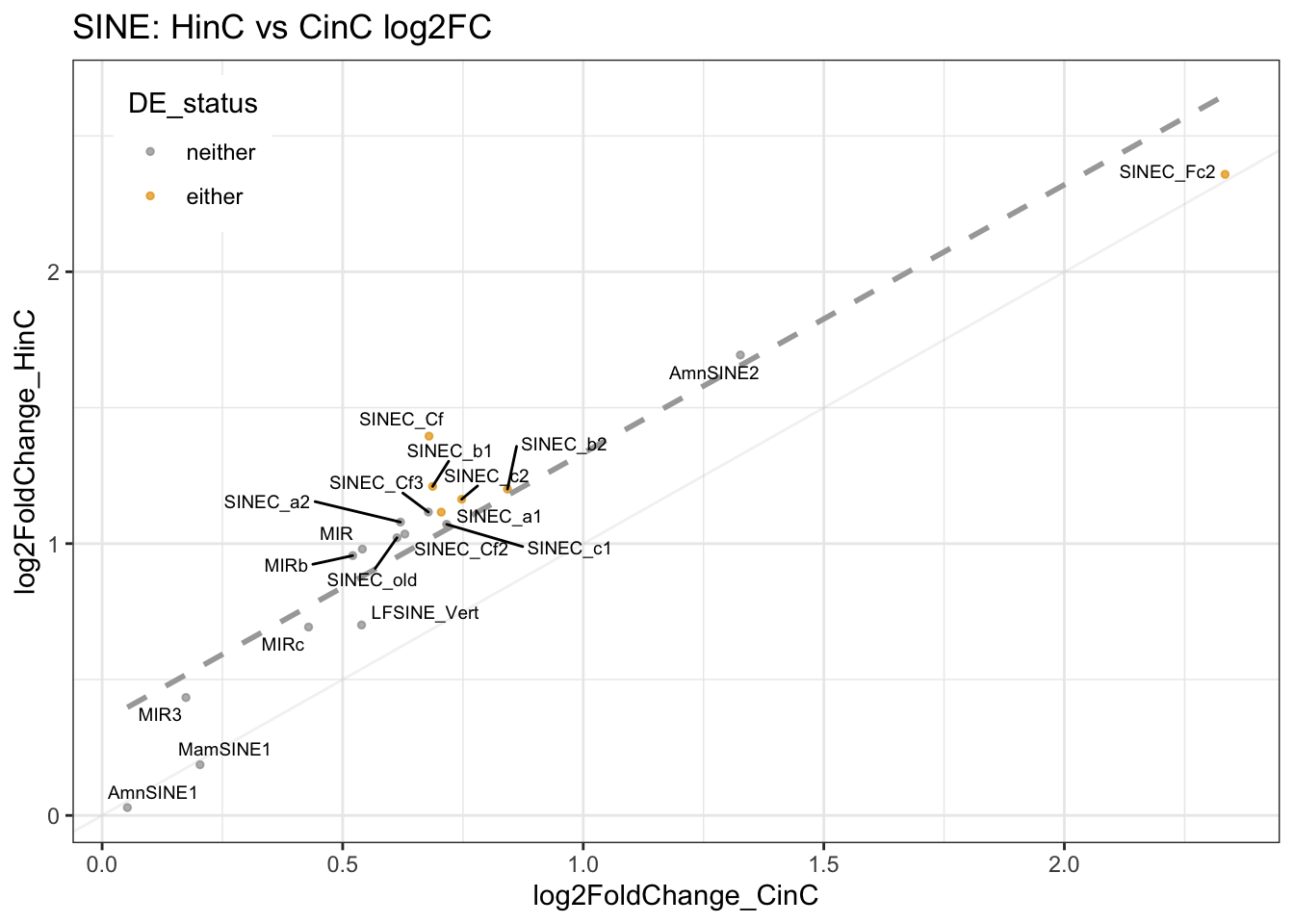
Figure 8.5: SINEs expression in HinC and CinC.
8.2.3 tRNA-Lys
Table below listed number of up-regulated repeats (name/group) in each SINE family:
SINE %>% dplyr::group_by(factor(repFamily_HinC)) %>%
dplyr::summarise(n = dplyr::n(),
HinC_sig = sum(padj_HinC < 0.05), CinC_sig=sum(padj_CinC < 0.05)) %>%
knitr::kable(caption="Number of up-regulated repeats (name/group) in each SINE family")## `summarise()` ungrouping output (override with `.groups` argument)| factor(repFamily_HinC) | n | HinC_sig | CinC_sig |
|---|---|---|---|
| Deu | 2 | 0 | 0 |
| MIR | 4 | 0 | 0 |
| tRNA | 2 | 0 | 0 |
| tRNA-Lys | 11 | 6 | 0 |
8.3 Satellites
Background: Satellites are consisted with short tandem repeats, and Canine satellite DNA may be more complicated and not restricted to the tandem repeats (Bentolila 1999). Based on RMAK, canine genome (canFam3) has 5 major satellites and satellite-like DNA - Bs, Carnivore Satellite (CarSat1, CarSat2), Satellite canis Familiaris (SAT1_CF, SAT2_CF, SAT3_CF, SAT4_CF and SAT6_CF), (CATTC)n, (GAATG)n and SUBTEL_sa. Bs, CarSat1, Carsat2, and SAT1_CF are the four satellites (repName) show some expression in DUX4 and DUXC expressed canine cell lines and therefore were analyzed. (CarSat1 and SAT1_CF are cetromeric satellite families.)
Results: Bs, CarSat1, and SAT1_CF were up-regulated in HinC. Bs was up-regulated in CinC; CarSat1 was moderately affected (\(padj = 0.14\)).
How does SAT1_CF or CarSat compare to HSETII and GSETII?
comb %>% dplyr::filter(repClass_HinC == "Satellite") %>%
dplyr::select(repName, log2FoldChange_HinC, log2FoldChange_CinC, padj_HinC, padj_CinC) %>%
knitr::kable(caption="Significant satallites in HinC and CinC transcriptome.")| repName | log2FoldChange_HinC | log2FoldChange_CinC | padj_HinC | padj_CinC |
|---|---|---|---|---|
| Bs | 6.906711 | 2.3038255 | 0.0000000 | 0.0002202 |
| CarSat1 | 7.476028 | 2.2907455 | 0.0000000 | 0.1413332 |
| Carsat2 | 1.315169 | 0.7970658 | 0.2892419 | 1.0000000 |
| SAT1_CF | 5.001051 | 1.3001238 | 0.0000000 | 0.4698271 |
# References
Sean C Shadle, Chao-Jen Wong, Sean R Bennett. 2019. “DUX4-Induced Bidirectional Hsatii Satellite Repeat Transcripts Form Intranuclear Double-Stranded Rna Foci in Human Cell Models of Fshd.” Human Molecular Genetics. https://doi.org/10.1093/hmg/ddz242.
Simone Bentolila, Jean-Louis Kessler, Jean-Marie Bach. 1999. “Analysis of Major Repetitive Dna Sequences in the Dog (Canis Familiaris) Genome.” Mammalian Genome.