Chapter 7 LTR activation
Previously @ref{repeats}, we showd the repeat elemnets profiling which revealed LTR activation by DUXC and DUX4 in the canine model. Here we investigated the subfamilies of LTR - ERVL and ERVL-MaLR. In the canine model, DUX4 and DUXC both activate elements form MaLR and ERVL. In addition, we found that DUXC prones to activate more ERVL elements (MLT2D, MER76, HERVL74, LTR1A2, LTR101, LTR33C) wheareas DUX4 to more MaLR elements (MLT1A/B/E2/J).
In this chapter, we (1) explain the background of ERVL and ERVL-MaLR subfamilies in human and murine, (2) exam ERVL and ERVL-MaLR activation by DUXC and DUX4 in canine myoblast, and (3) compare to the LTR subfamily activation in human myoblast.
Load and datsets and libraries
# define directory
pkg_dir <- "~/CompBio/canFam3.DuxFamily"
fig_dir <- file.path(pkg_dir, "figures")
data_dir <- file.path(pkg_dir, "data")
# load library
suppressPackageStartupMessages(library(DESeq2))
suppressPackageStartupMessages(library(tidyverse))
suppressPackageStartupMessages(library(tidyr))
suppressPackageStartupMessages(library(ggrepel))
suppressPackageStartupMessages(library(purrr))
suppressPackageStartupMessages(library(canFam3.rmsk))
load(file.path("~/CompBio/hg38/hg38.rmsk.rda"))
suppressPackageStartupMessages(library(wesanderson)) # modern palettes
# load datasets and local functions
source(file.path(pkg_dir, "scripts", "tools.R"))
load(file.path(pkg_dir, "data", "HinC_rmsk.rda")) # made in chapter 6
load(file.path(pkg_dir, "data", "CinC_rmsk.rda")) # made in chapter 6
load(file.path(pkg_dir, "data", "CALTinC_rmsk.rda")) # made in chapter 6
load(file.path(pkg_dir, "data", "HinH_rmsk.rda"))7.1 Takaway
The ERVL-MaLR elements in canine are categorized as MLT1, presenting ancient mammalian LTRs. Most of the MLT1s annotated in the canine genome were mobilized before the split of common ancient mammalian of canine and primate lineages, except MLT1A0 (-int) and MLT1A1 (-int) that might have become more mobilized in the primate lineage. Compared with DUXC, DUX4 induces higher expression levels of MLT1s in canine, though its affinity toward MLT1s has no apparent correlation with the copy number or mobilization in the primate lineage.
ERVL1: ERV1 (denoted LTR* and MER*) are enriched in both DUX4 and DUXC expressed canine skeletal muscle; 50% and 18% of ERVL1 elements are positively affected by DUXC and DUXC, respectively. DUX4 induced more carnivore-specific ERV1 (CarLTR, CfERV1, CdERVF) than DUXC.
ERVL-MaLR: Studies indicate that DUX4 uses MLT1 as promoter to re-program nearby genes’s structure during cleavage-stage and FSHD affected skeletal muscle. MLT1 presents ancient mammalian LTRs whereas THE1 and MST radiats in primate lineage. In canine genome, most of the ERVL-MaLR elements are MLT1 and mobilized before the primate lineages; the copy numbers of MLT1s existed in canine genome are compariable to that of in human genome, excpet MLT1A1 (2.2) and MLT1A0-int (>100), both of which are DUX4 preferred repeats for retrontransposon-mediated transcriptome in humand and canine myoblosts. DUX4 and DUXC positively affect 51% and 25% of ERVLs, respectively.
ERVL: (denoted MLT2, MER* and LTR*) DUX4 and DUXC positively affect 49% and 23% of ERVLs, respectively.
7.2 background in murine and human
Though sharing common ancestry, MuERV-L (denoted MT2_Mm and MT2) evolved differntly from MaLR in murine. The ERVL-MaLR include three major subfamilies: MLT, ORR1 and MT. MLT; MLT1 and MLT2, present ancenstral mammalian LTR; ORR1 and MT are the youngest repeats in rodent, and they radiate during rodent evolution. (Note: MT, ORR1 nd MT2 radiate during rodent evolution.)
Whidden et al. (Jennifer L. Whiddon 2017), Hackdrison et al. (???) and Franke et al (Vedran Franke 2017) have investigated the activities of the mouse endogenous retrovirus type-L (MuERV-L) and nonautonomous mammalian (ERVL-MaLR) subfamilies under the influence of Dux and early embroynic activation. The studies confirm the species-specific preference on activation of retrotransposon subfamilies during the 2C-like (cleavage) stage or Dux/DUX4 over-expression: human bound to ancestral (MLT1) and primate-specific (THE1, MST) ERVL-MaLR subfamilies whereas murine to MuERV-L and young submailies of MaLR that evolved during rodent evolution.
Frank 2015 (Vedran Franke 2017) showed that one of the ERVL functions is gene-remodeling via four co-options:
1. 5’ exon via retrotransoson-derived spliced donor (SD)
2. 3’ exon via retrotransposon-derived SD or poly-(A)
3. internal exon via retrotransposon-derived SD or DA
4. intraexonic, not contribute to mRNA process
In general, the younger ERVL subfamilies (Vedran Franke 2017) - ORR1, MT, MT2 - showed higher propotion of 5’exon co-option than older family (MLT) in murine. Human bound to use more MLT1 (subfamily of MaLR) 5’exon co-option, contributing MLT to promotor and first exons. Unlike rodent, human does not use MT2 for 5’exon co-option; Rodent uses more MT (subfamily of MaLR) and MT2 (subfamily of MuERL-V) 5’exon co-option. Whidden et al. (Jennifer L. Whiddon 2017) confirmed that Dux and DUX4 express different subsets of MaLR transcripts in C2C12: Dux uses MT2 (MuERL-V) and MT as promoters whereas DUX4 uses MLT as promoters to activate the targets.
Previous chapter @ref{repeats} stated that DUXC showed higher propotion of ERVL differential expression whereas DUX4 showed higher propotion in that of MaLR. Althought we observe differntial expression in MaLR in DUXC and DUX4, the question is are they in distinct subfamilies of MaLR?
7.3 ERVL subfamily activation in canine
# extract ERVL retroelement
duxc_ERVL <- CinC_rmsk$res_df %>%
dplyr::filter(repFamily %in% c("ERVL", "ERVL?")) %>%
dplyr::mutate(up_reg = ifelse(padj < 0.05 & log2FoldChange >0, TRUE, FALSE)) %>%
dplyr::mutate(up_reg = replace(up_reg, is.na(up_reg), FALSE))
dux4_ERVL <- HinC_rmsk$res_df %>%
dplyr::filter(repFamily %in% c("ERVL","ERVL?")) %>%
dplyr::mutate(up_reg = ifelse(padj < 0.05 & log2FoldChange > 0, TRUE, FALSE)) %>%
dplyr::mutate(up_reg = replace(up_reg, is.na(up_reg), FALSE))
# combine dux4 and duxc and make scatter plot; label DUX4 and DUX4 only
ERVL <- full_join(duxc_ERVL, dux4_ERVL, by="repName", suffix=c("_DUXC", "_DUX4")) %>%
dplyr::mutate(status = "none") %>%
dplyr::mutate(status = replace(status, padj_DUX4 < 0.05, "DUX4")) %>%
dplyr::mutate(status = replace(status, padj_DUXC < 0.05, "DUXC")) %>%
dplyr::mutate(status = replace(status, padj_DUXC < 0.05 & padj_DUX4 < 0.05, "both")) %>%
dplyr::mutate(status = factor(status, levels=c("none","DUX4", "DUXC", "both"))) %>%
dplyr::mutate(show_name = as.character(repName)) %>%
dplyr::mutate(show_name = ifelse(status %in% c("DUX4", "DUXC", "both"), show_name, ""))my_color <- c("#999999",
as.character(wes_palette(n=5, name="FantasticFox1", type="discrete"))[3:5])
gg1 <- ggplot(ERVL, aes(x=log2FoldChange_DUXC, y=log2FoldChange_DUX4, color=status)) +
geom_point(size=1) +
scale_color_manual(values=my_color) +
#geom_smooth(method="lm", se=FALSE, show.legend=FALSE, color="gray66", alpha=0.5,
# linetype="dashed") +
geom_abline(intercept=0, slope=1, color="gray75", alpha=0.2, size=0.5) +
labs(title="ERVL: HinC vs CinC log2FC") +
theme_bw()+
theme(legend.position = c(0.1, 0.83)) +
geom_text_repel(aes(label=show_name), size=2.5, show.legend=FALSE)
gg1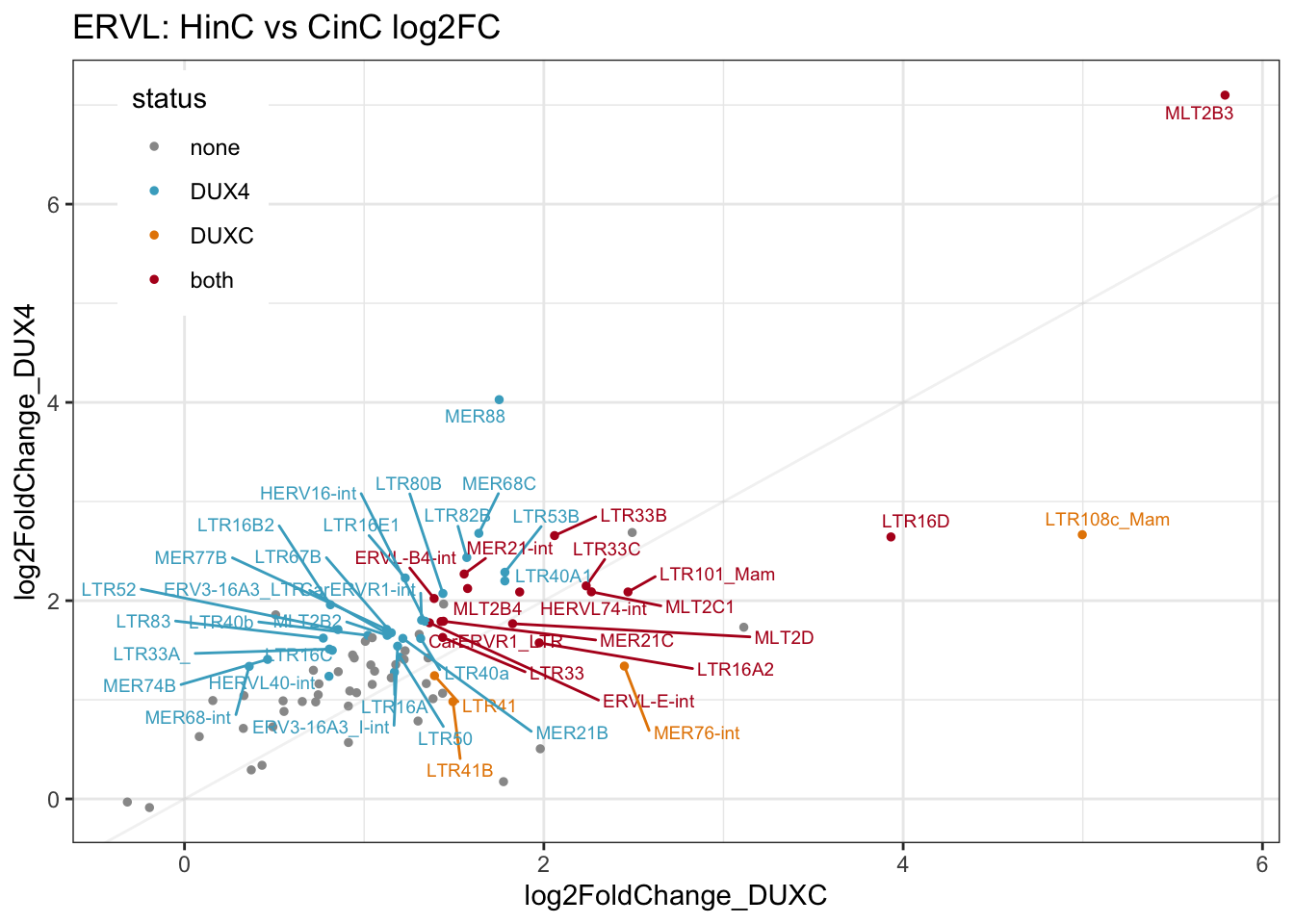
Figure 7.1: Scatter plot of log2FC of DUXC and DUX4
7.4 MaLR activation in canine
# extract ERVL subfamilies
duxc_MaLR <- CinC_rmsk$res_df %>%
dplyr::filter(repFamily == "ERVL-MaLR") %>%
dplyr::mutate(up_reg = ifelse(padj < 0.05 & log2FoldChange >0, TRUE, FALSE)) %>%
dplyr::mutate(up_reg = replace(up_reg, is.na(up_reg), FALSE))
dux4_MaLR <- HinC_rmsk$res_df %>%
dplyr::filter(repFamily == "ERVL-MaLR") %>%
dplyr::mutate(up_reg = ifelse(padj < 0.05 & log2FoldChange > 0, TRUE, FALSE)) %>%
dplyr::mutate(up_reg = replace(up_reg, is.na(up_reg), FALSE))
# combine dux4 and duxc and make scatter plot; label DUX4 and DUX4 only
MaLR <- full_join(duxc_MaLR, dux4_MaLR, by="repName", suffix=c("_DUXC", "_DUX4")) %>%
dplyr::mutate(status = "none") %>%
dplyr::mutate(status = replace(status, padj_DUX4 < 0.05, "DUX4")) %>%
dplyr::mutate(status = replace(status, padj_DUXC < 0.05, "DUXC")) %>%
dplyr::mutate(status = replace(status, padj_DUXC < 0.05 & padj_DUX4 < 0.05, "both")) %>%
dplyr::mutate(status = factor(status, levels=c("none","DUX4", "DUXC", "both"))) %>%
dplyr::mutate(show_name = as.character(repName)) %>%
dplyr::mutate(show_name = ifelse(status %in% c("DUX4", "DUXC", "both"), show_name, ""))my_color <- c("#999999",
as.character(wes_palette(n=5, name="FantasticFox1", type="discrete"))[3:5])
gg1 <- ggplot(MaLR, aes(x=log2FoldChange_DUXC, y=log2FoldChange_DUX4, color=status)) +
geom_point(size=1) +
scale_color_manual(values=my_color) +
#geom_smooth(method="lm", se=FALSE, show.legend=FALSE, color="gray66", alpha=0.5,
# linetype="dashed") +
geom_abline(intercept=0, slope=1, color="gray75", alpha=0.2, size=0.5) +
labs(title="ERVL-MaLR: HinC vs CinC log2FC") +
theme_bw()+
theme(legend.position = c(0.1, 0.83)) +
geom_text_repel(aes(label=show_name), size=2.5, show.legend=FALSE)
gg1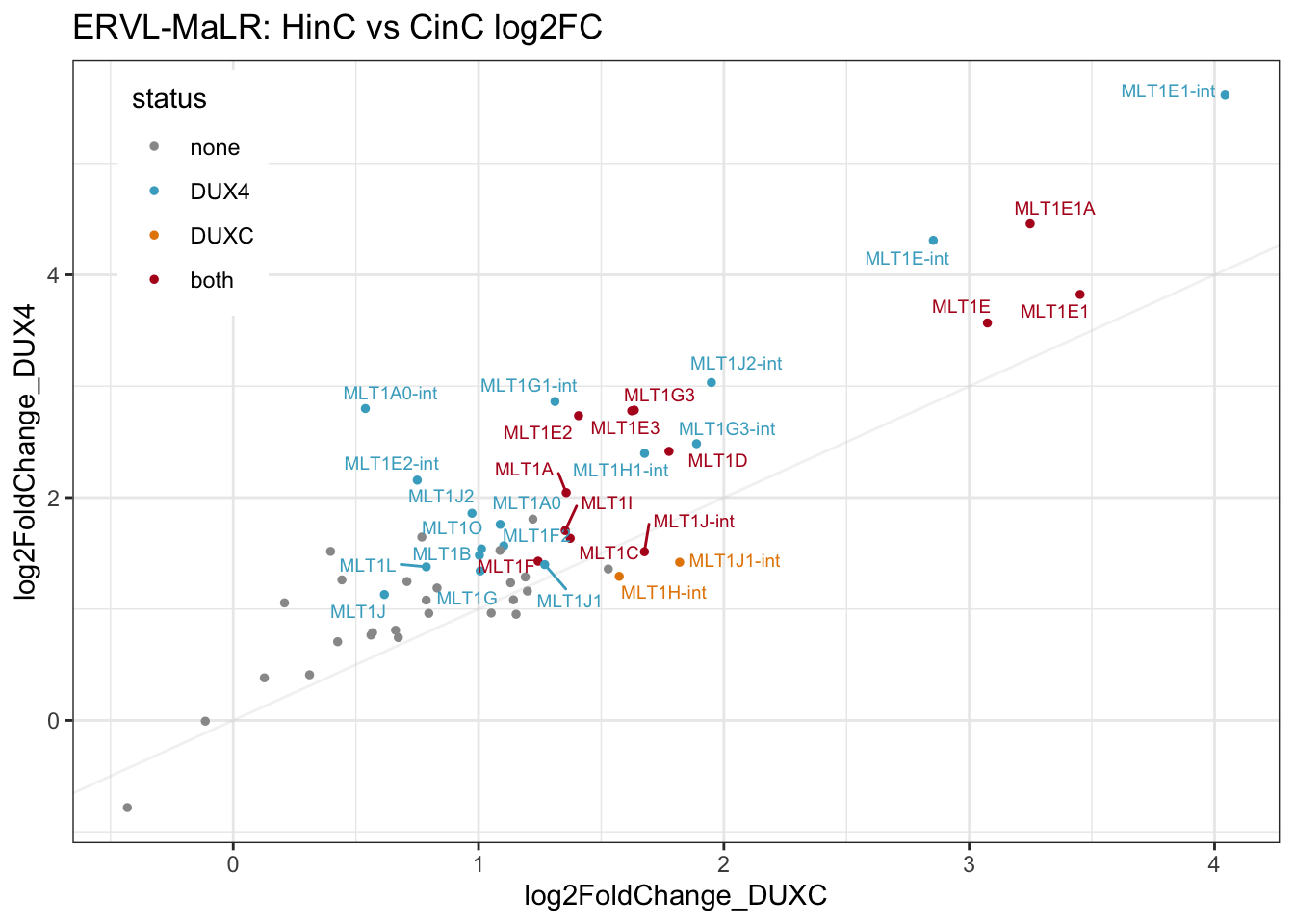
Figure 7.2: Scatter plot of log2FC of DUXC and DUX4
Visualizaiton of the number of ERVL subfamilies activated by DUX4 and DUXC in canine.
7.5 Compare to CinC and HinC to HinH
Here we compared LTR retroelment activation of CinC and HinC to that of HinH. We aimed to answer whether copy number is associated with higher induction by DUX4 than by DUXC? Update: The answer is no. Copy number is not associated with higher induction of DUX4 nor DUXC.
7.5.1 Copy number
The code below estimates the copy number of LTR subfamilies in hg38 and canFam3.
# get the number of instance of a repeat element within a family
get_family_copy_number <- function(hg38.rmsk, canFam3.rmsk, family_name) {
if (!any(c(levels(hg38.rmsk$repFamily), levels(hg38.rmsk$repFamilf)) %in% family_name))
stop(paste(family_name, sep=", "), " not found in both hg38 and canFam3 rmsk!")
hg38_sub<- plyranges::filter(hg38.rmsk, repFamily %in% family_name) %>%
plyranges::mutate(repClass = factor(repClass), repFamily = factor(repFamily),
repName = factor(repName))
hg38_copy_n <- as(mcols(hg38_sub), "data.frame") %>% dplyr::count(repName)
# canine
canFam3_sub <- plyranges::filter(canFam3.rmsk, repFamily %in% family_name) %>%
plyranges::mutate(repClass = factor(repClass), repFamily = factor(repFamily),
repName = factor(repName))
canFam3_copy_n <- as(mcols(canFam3_sub), "data.frame") %>% dplyr::count(repName)
full_join(canFam3_copy_n, hg38_copy_n,
by="repName", suffix=c(".canFam3", ".hg38")) %>%
dplyr::mutate(ratio = n.hg38 / n.canFam3)
}
# estimate copy number of MaLR, ERV1 and ERVL retroelemtns in canine and human using RMSK
MaLR_copy_number <- get_family_copy_number(hg38.rmsk, canFam3.rmsk, "ERVL-MaLR")
ERVL_copy_number <- get_family_copy_number(hg38.rmsk, canFam3.rmsk, "ERVL")
ERV1_copy_number <- get_family_copy_number(hg38.rmsk, canFam3.rmsk, "ERV1")
# HERV: expended in human/primate lineage
head(MaLR_copy_number)## repName n.canFam3 n.hg38 ratio
## 1 MLT-int 171 394 2.304094
## 2 MLT1-int 226 251 1.110619
## 3 MLT1A 8364 9312 1.113343
## 4 MLT1A-int 371 456 1.229111
## 5 MLT1A0 11780 21235 1.802632
## 6 MLT1A0-int 414 971 2.345411Tidy up activated LTR retroelements MaLR, ERVL, and ERV1) in HinC, CinC and HinH together.
# 1. union the activated LTR retroelements of HinC and CinC; then append HinH's statistics
# 2. incorporate with copy number
activated_LTRs <- inner_join(HinC_rmsk$res_df, CinC_rmsk$res_df,
by="repName", suffix=c("_HinC", "_CinC")) %>%
left_join(dplyr::select(HinH_rmsk$res_df, repName, log2FoldChange, padj), by="repName") %>%
rename(repFamily = repFamily_CinC, log2FoldChange_HinH = log2FoldChange, padj_HinH = padj) %>%
dplyr::filter(repFamily %in% c("ERVL-MaLR", "ERVL", "ERV1")) %>%
dplyr::filter(log2FoldChange_HinC > 0 | log2FoldChange_CinC > 0 | log2FoldChange_HinH > 0) %>%
dplyr::filter(padj_HinC < 0.05 | padj_CinC < 0.05 | padj_HinH < 0.05) %>%
dplyr::select(repName, repFamily,
log2FoldChange_HinC, padj_HinC, log2FoldChange_CinC, padj_CinC,
log2FoldChange_HinH, padj_HinH)
logfc_gather <- gather(activated_LTRs, key=tr_factor, value=logFC,
log2FoldChange_HinC, log2FoldChange_CinC, log2FoldChange_HinH,
-repName, -repFamily) %>%
dplyr::mutate(padj = padj_HinC) %>%
dplyr::mutate(padj = ifelse(tr_factor == "log2FoldChange_CinC", padj_CinC, padj)) %>%
dplyr::mutate(padj = ifelse(tr_factor == "log2FoldChange_HinH", padj_HinH, padj)) %>%
dplyr::select(repName, repFamily, tr_factor, logFC, padj) %>%
dplyr::mutate(sig = ifelse(padj < 0.05, TRUE, FALSE)) %>%
dplyr::mutate(tr_factor = sapply(strsplit(tr_factor, "_", fixed=TRUE), "[[", 2))7.5.2 ERVL-MaLR
The bar plot below displays signicantly expressed ERVL-MaLR elements in either CinC, HinC or HinH (\(adj-p < 0.05\)). Only MLT1A1 and MLT1A0-int have higher copy number in human (ratio > 2) compared to canine (ratio > 0), but they don’t have higher induction in human.
data <- dplyr::filter(logfc_gather, repFamily == "ERVL-MaLR") %>%
left_join(MaLR_copy_number, by="repName") %>%
dplyr::mutate(repName = ifelse(ratio > 2, paste0(repName, "*"), repName))
gg1 <- ggplot(data, aes(x = repName, y=logFC, fill=sig)) +
geom_bar(stat="identity") +
facet_wrap ( ~ tr_factor) +
coord_flip() +
theme_minimal() +
labs(title="ERVL-MaLR") +
scale_fill_manual(values=c("#999999", "#E69F00")) +
theme(legend.position = "none")
gg1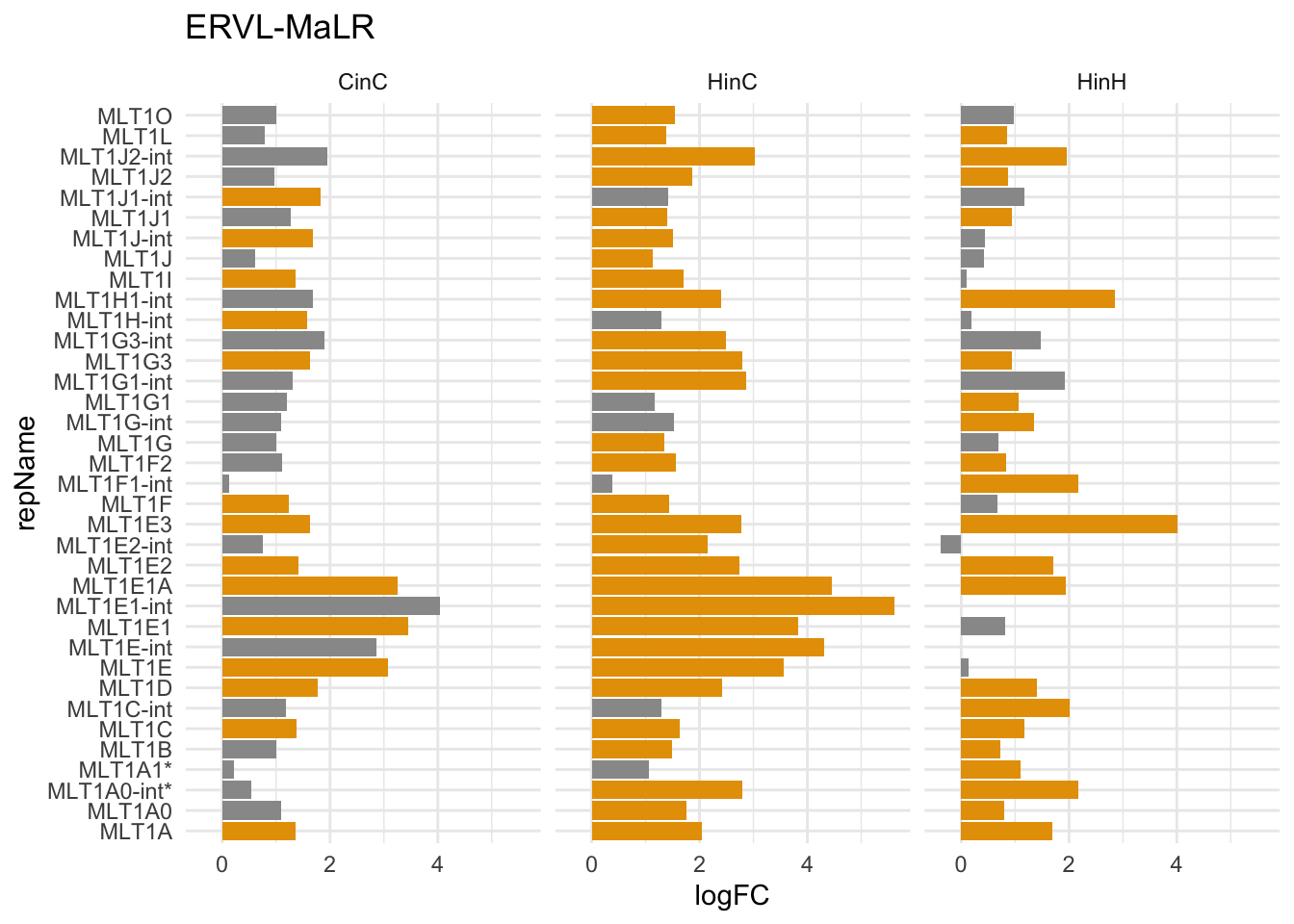
Figure 7.3: repName expression (logFC) in ERVL-MaLR family in CinC, HinC and HinH; Orange indicates up/down-regulation; * indicates higher copy number in human.
7.5.3 ERVL
Shown below is the significently expressed ERVL elememts expression in either CinC, HinC, or HinH. MER21B and MER21-int have highr copy number in human.
data <- dplyr::filter(logfc_gather, repFamily == "ERVL") %>%
left_join(ERVL_copy_number, by="repName") %>%
dplyr::mutate(repName = ifelse(ratio > 2, paste0(repName, "*"), repName))
gg2 <- ggplot(data, aes(x = repName, y=logFC, fill=sig)) +
geom_bar(stat="identity") +
facet_wrap ( ~ tr_factor) +
coord_flip() +
theme_minimal() +
labs(title="ERVL") +
scale_fill_manual(values=c("#999999", "#E69F00")) +
theme(legend.position = "none")
gg2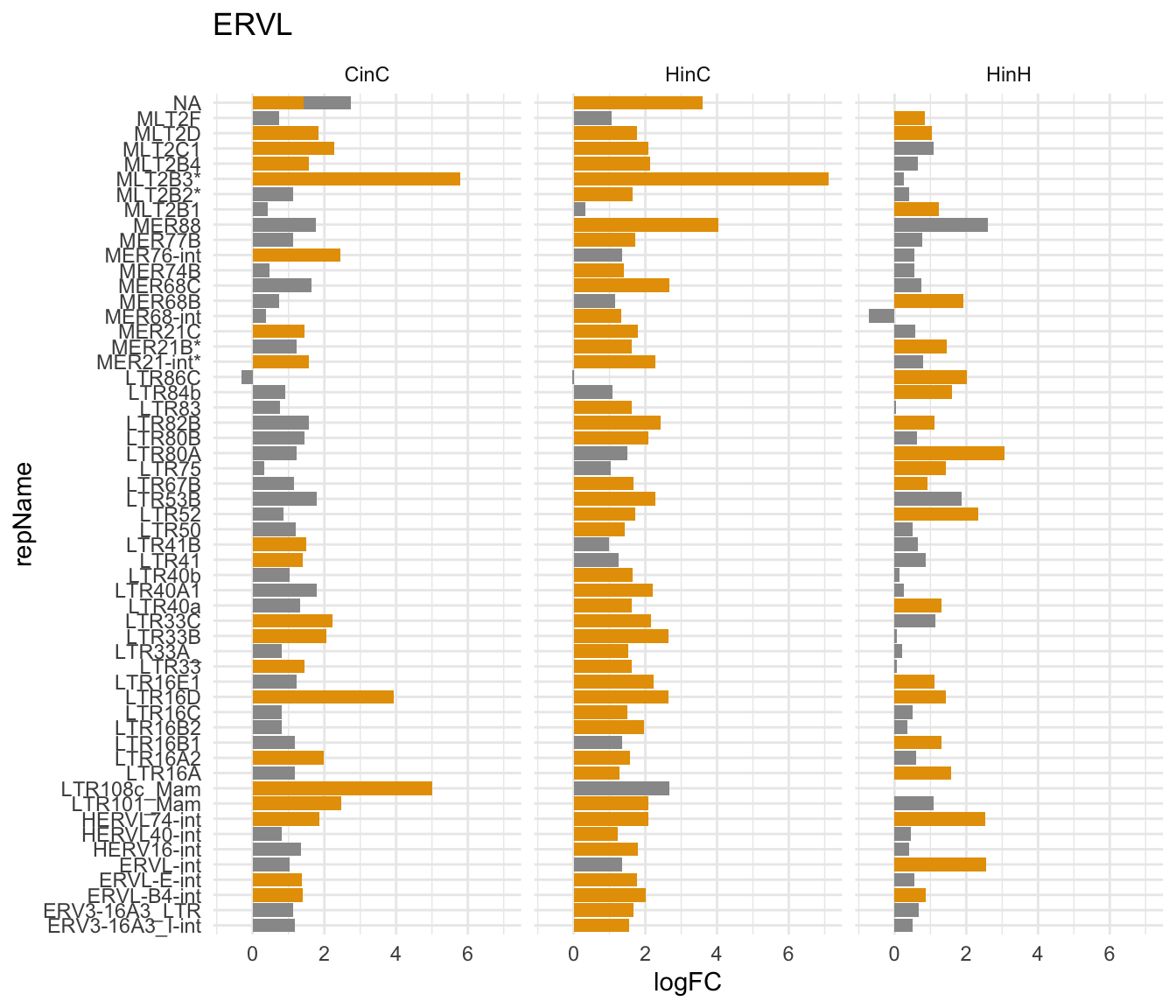
Figure 7.4: repName expression (logFC) in ERVL family in CinC, HinC and HinH; Orange indicates up/down-regulation; * indicates higher copy number in human.
7.5.4 ERV1
Shown below is the significently expressed ERV1 elements in either CinC, HinC, or HinH. Suprisingly, DUX4 up-regulated more carnior-specific retroelement than DUXC in canine.
data <- dplyr::filter(logfc_gather, repFamily == "ERV1") %>%
left_join(ERV1_copy_number, by="repName") %>%
dplyr::mutate(repName = ifelse(!is.na(ratio) & ratio > 2, paste0(repName, "*"), repName)) %>%
dplyr::mutate(repName = ifelse(is.na(ratio), paste0(repName, "#"), repName))
gg3 <- ggplot(data, aes(x = repName, y=logFC, fill=sig)) +
geom_bar(stat="identity") +
facet_wrap ( ~ tr_factor) +
coord_flip() +
theme_minimal() +
labs(title="ERV1") +
scale_fill_manual(values=c("#999999", "#E69F00")) +
theme(legend.position = "none")
gg3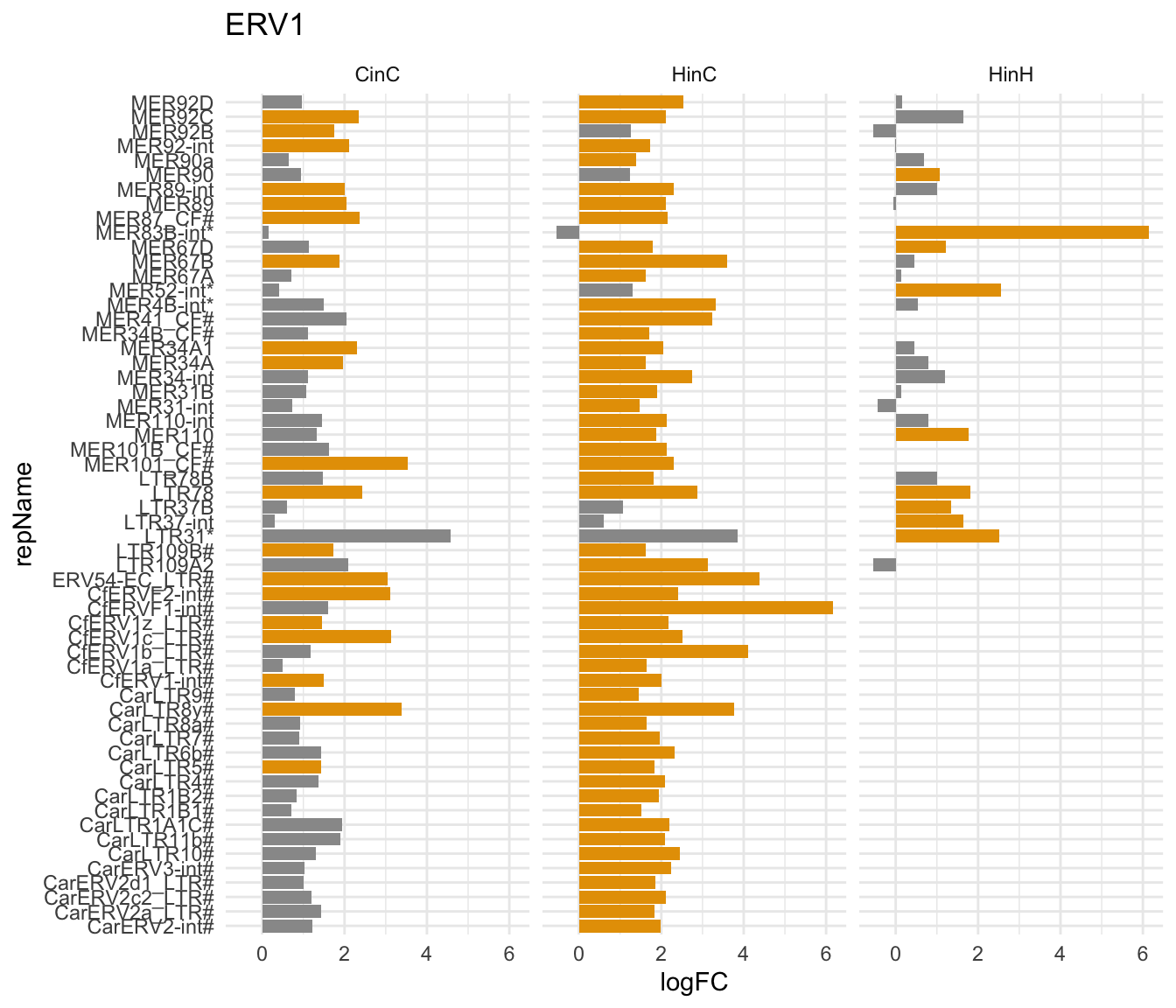
Figure 7.5: repName expression (logFC) in ERV1 family in CinC, HinC and HinH; Orange indicates up/down-regulation; * indicates higher copy number in human. # indicates the copy number ratio (human/canine) is not available.
7.6 DUXC-ALT and ERVL transcriptions
Does DUXC-ALT induce ERVL expression? There are 683 repeat elements exhibiting some expression in DUXC-ALT expressed cell line. The binding sites of DUXC-ALT suggested that DUXC-ALT mostly binds to LTR retroelements. We aim to exam further into DUXC-ALT’s function by looking at the expession fold changes.
7.6.1 ERV subfamily expression
Shown below is the histogram of DUXC-ALT’s repeat elements expression. Most the expression of the expression of repeat elements do not have apparent changes by DUXC-ALT, however, some ERV transcripts are present with mild fold changes. This histogram below shows that a few repeat element transcripts are present with mild fold changes.
calt_df <- CALTinC_rmsk$res_df
ggplot(calt_df, aes(x=log2FoldChange)) +
geom_histogram(bins=30, color="white", fill="#DD8D29") +
theme_bw() +
labs(title="Repeat element expression in DUXC-ALT") +
coord_cartesian(xlim=c(-2,2)) +
theme(plot.title = element_text(size=10))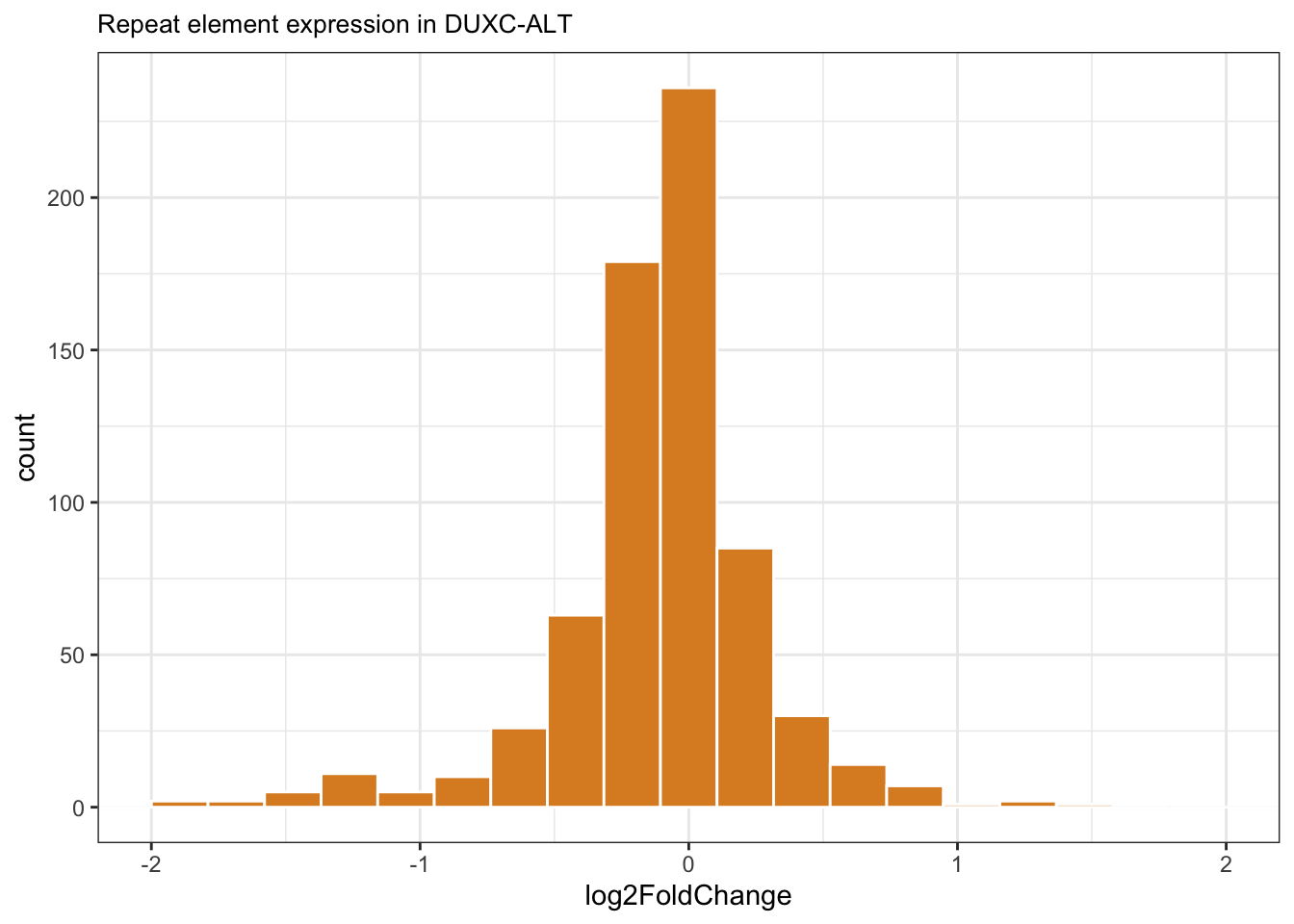
Figure 7.6: Histogram of log2 Fold Change of CALTinC repeat elements
Disregard the p-adj and p-value, we listed the repeat elements with log2 fold change greater and less then 0.5, and most of them are ERVL1. This might support the peak calling of DUXC-Alt where more than 60% of binding sites are overlapping with LTRs.
# generate table of repeat element with log2FC > 0.5
calt_up <- calt_df %>% dplyr::filter(log2FoldChange > 0.5)
calt_up %>% dplyr::filter(repFamily %in% c("ERV1", "ERVL?", "ERVL", "ERVL-MaLR", "Gypsy", "LTR")) %>%
dplyr::select(-baseMean, -lfcSE, -stat) %>%
knitr::kable(caption="Repeat element with log2FC > 0.5 in DUXC-Alt.")| repName | log2FoldChange | pvalue | padj | repClass | repFamily |
|---|---|---|---|---|---|
| CarERV2b1_LTR | 0.7129453 | 1 | 1 | LTR | ERV1 |
| CarLTR1A1C | 0.6510292 | 1 | 1 | LTR | ERV1 |
| LTR109A2 | 0.8342682 | 1 | 1 | LTR | ERV1 |
| LTR55 | 0.6147695 | 1 | 1 | LTR | ERVL? |
| LTR72B_CF | 0.6561270 | 1 | 1 | LTR | ERV1 |
| LTR81B | 0.5263868 | 1 | 1 | LTR | Gypsy |
| LTR90B | 0.9274856 | 1 | 1 | LTR | LTR |
| MamGypLTR3 | 0.7477494 | 1 | 1 | LTR | Gypsy |
| MamGypLTR3a | 0.6591307 | 1 | 1 | LTR | Gypsy |
| MER101_CF | 0.5369590 | 1 | 1 | LTR | ERV1 |
| MER88 | 0.8910420 | 1 | 1 | LTR | ERVL |
| MLT2E | 0.6513342 | 1 | 1 | LTR | ERVL |
# generate table of repeat elements with log2FC < -1
calt_down <- calt_df %>% dplyr::filter(log2FoldChange < -1)
calt_down %>%
dplyr::select(-baseMean, -lfcSE, -stat) %>%
knitr::kable(caption="Repeat element with log2FC < -1 in DUXC-Alt.")| repName | log2FoldChange | pvalue | padj | repClass | repFamily |
|---|---|---|---|---|---|
| (CATCG)n | -1.378227 | 0.8176728 | 1 | Simple_repeat | Simple_repeat |
| (CCCCAA)n | -1.329845 | 0.8430315 | 1 | Simple_repeat | Simple_repeat |
| (CCGGA)n | -1.430167 | 0.7516265 | 1 | Simple_repeat | Simple_repeat |
| (GAGAA)n | -3.657527 | 0.4001370 | 1 | Simple_repeat | Simple_repeat |
| (GTGTG)n | -1.087761 | 0.9440174 | 1 | Simple_repeat | Simple_repeat |
| (TAGG)n | -2.614182 | 0.3792413 | 1 | Simple_repeat | Simple_repeat |
| (TTCGGG)n | -1.300816 | 0.4825708 | 1 | Simple_repeat | Simple_repeat |
| 5S | -1.405084 | 0.6721832 | 1 | rRNA | rRNA |
| 7SLRNA | -1.598665 | 0.0323175 | 1 | srpRNA | srpRNA |
| CarLTR1A2B | -1.363858 | 0.7539420 | 1 | LTR | ERV1 |
| CfERV1b_LTR | -1.194750 | 0.7512672 | 1 | LTR | ERV1 |
| CfERVF1_LTR | -1.222406 | 0.7152418 | 1 | LTR | ERV1 |
| DNA1_Mam | -1.104721 | 0.9054235 | 1 | DNA | TcMar |
| Eulor5A | -1.665583 | 0.6233808 | 1 | DNA? | DNA? |
| L1M3b | -1.215819 | 0.6528945 | 1 | LINE | L1 |
| L1M3e | -1.278423 | 0.6166096 | 1 | LINE | L1 |
| LTR109A1 | -1.536070 | 0.6485770 | 1 | LTR | ERV1 |
| LTR81C | -1.375129 | 0.5548298 | 1 | LTR | Gypsy |
| LTR91 | -1.879468 | 0.5683125 | 1 | LTR | ERVL |
| MamGypLTR2c | -1.111055 | 0.7839170 | 1 | LTR | Gypsy |
| MER45R | -1.272985 | 0.6944189 | 1 | DNA | hAT-Tip100 |
| MER74C | -1.256894 | 0.7651719 | 1 | LTR | ERVL |
| MER87 | -1.367775 | 0.8284964 | 1 | LTR | ERV1 |
| MLT1A0-int | -1.083928 | 0.8646201 | 1 | LTR | ERVL-MaLR |
| MLT1A1 | -1.892689 | 0.5746772 | 1 | LTR | ERVL-MaLR |
| MLT1G1-int | -1.273883 | 0.5981955 | 1 | LTR | ERVL-MaLR |
DUXC-Alt suppressed many of ERVL and ERVL-MaLR (MLT1) elements, though weak p-values are presented.
# References
Vedran Franke, Rosa Karlic, Sravya Ganesh. 2017. “Long Terminal Repeats Power Evolution of Genes and Gene Expression Programs in Mammalian Oocytes and Zygotes.” Genomic Research, no. 27. https://doi.org/10.1101/gr.216150.116.