11 DNA methylation of FSHD D4Z4 allele
We conducted bisulfite sequencing on bilateral TA muscle biopsies from 34 subjects. In this chapter, we present the following:
- The CpG methylation percentages for subjects with the 4qA161S and 4qA161L alleles.
- The correlation of methylation levels between left and right TA biopsies.
- The association between methylation and STIR status.
- Correlations with other variables, including muscle strength, disease molecular signatures, whole muscle fat percentage, and clinical severity score (CSS).
Note: We excluded subject 01-0022 in this analysis due to its duplication in D4Z4 repeat regions.
11.1 Loading and cleaning data
We load two data sets from the data directory:
1. comprehensive_df.rda: clinical and MRI characteristic data
2. bs_methyl.rda: DNA methylation data
library(tidyverse)
library(corrr)
library(kableExtra)
pkg_dir <- "/Users/cwon2/CompBio/Wellstone_BiLateral_Biopsy"
load(file.path(pkg_dir, "data", "bs_methyl.rda"))
load(file.path(pkg_dir, "data", "comprehensive_df.rda"))
comprehensive_df <- comprehensive_df %>%
dplyr::rename_with(~str_remove(., '-logSum'), ends_with('-logSum'))Tidy up the bisulfite sequencing data.
bss <- bs_methyl %>%
dplyr::mutate(`4A161S` = if_else(grepl("4A161S", `haplotype_chr4`),
TRUE, FALSE)) %>%
dplyr::mutate(`4A161L` = if_else(grepl("4A161L", `haplotype_chr4`),
TRUE, FALSE)) %>%
dplyr::filter(! Subject == "01-0022") %>%
dplyr::mutate(allele=if_else(`use BSSL`, "4qA161L", "4qA161S")) %>%
dplyr::mutate(allele=factor(allele, levels=c("4qA161S", "4qA161L")))| Subject | location | sample_id | allele | BSS Value | Predicted Haplotype |
|---|---|---|---|---|---|
| 01-0019 | R | 01-0019R | 4qA161S | 8.000000 | 4A161S(5RU)/4B163(32RU), 10A164/10A164 |
| 01-0019 | L | 01-0019L | 4qA161S | 4.000000 | 4A161S(5RU)/4B163(32RU), 10A164/10A164 |
| 01-0020 | R | 01-0020R | 4qA161S | 4.000000 | 4A161S(5RU)/4A161S(22RU), 10A166/10A166 |
| 01-0020 | L | 01-0020L | 4qA161S | 4.000000 | 4A161S(5RU)/4A161S(22RU), 10A166/10A166 |
| 01-0021 | R | 01-0021R | 4qA161L | 14.285710 | 4A161L(10RU)/4B163(16RU), 10A166/10A166 |
| 01-0021 | L | 01-0021L | 4qA161L | 19.047620 | 4A161L(10RU)/4B163(16RU), 10A166/10A166 |
| 01-0023 | R | 01-0023R | 4qA161S | 8.000000 | 4A161S/4B163, 10A166/10A166 |
| 01-0023 | L | 01-0023L | 4qA161S | 8.000000 | 4A161S/4B163, 10A166/10A166 |
| 01-0024 | R | 01-0024R | 4qA161S | 8.000000 | 4A161S(7RU)/4B163(17RU), 10A166/10A166 |
| 01-0024 | L | 01-0024L | 4qA161S | 4.000000 | 4A161S(7RU)/4B163(17RU), 10A166/10A166 |
| 01-0025 | R | 01-0025R | 4qA161S | 4.000000 | 4A161S(6RU)/4A161S(16RU), 10A166/10A166 |
| 01-0025 | L | 01-0025L | 4qA161S | 4.000000 | 4A161S(6RU)/4A161S(16RU), 10A166/10A166 |
| 01-0026 | R | 01-0026R | 4qA161S | 0.000000 | 4A161S(6RU)/4A161L(45RU), 10A166/10A168 |
| 01-0026 | L | 01-0026L | 4qA161S | 0.000000 | 4A161S(6RU)/4A161L(45RU), 10A166/10A168 |
| 01-0028 | R | 01-0028R | 4qA161S | 8.000000 | 4A161S(7RU)/4A161S(33RU), 10A166/4A166 |
| 01-0028 | L | 01-0028L | 4qA161S | 8.000000 | 4A161S(7RU)/4A161S(33RU), 10A166/4A166 |
| 01-0029 | R | 01-0029R | 4qA161S | 4.000000 | 4A161S(7RU?)/4A161S, 10A166/4A166 |
| 01-0029 | L | 01-0029L | 4qA161S | 0.000000 | 4A161S(7RU?)/4A161S, 10A166/4A166 |
| 01-0030 | R | 01-0030R | 4qA161S | 4.166667 | 4A161S(5RU)/4B168(31RU), 10A166/4A166 |
| 01-0030 | L | 01-0030L | 4qA161S | 12.000000 | 4A161S(5RU)/4B168(31RU), 10A166/4A166 |
| 01-0031 | R | 01-0031R | 4qA161S | 12.000000 | 4A161S(7RU)/4B163(34RU), 10A166/10A166 |
| 01-0031 | L | 01-0031L | 4qA161S | 8.000000 | 4A161S(7RU)/4B163(34RU), 10A166/10A166 |
| 13-0001 | R | 13-0001R | 4qA161S | 4.000000 | 4A161S(4RU)/4A161L(16RU:52RU=1:1), 10A166/10B161T |
| 13-0001 | L | 13-0001L | 4qA161S | 0.000000 | 4A161S(4RU)/4A161L(16RU:52RU=1:1), 10A166/10B161T |
| 13-0002 | R | 13-0002R | 4qA161S | 4.000000 | 4A161S(5RU)/4B163(28RU), 10A166/10A166 |
| 13-0002 | L | 13-0002L | 4qA161S | 4.000000 | 4A161S(5RU)/4B163(28RU), 10A166/10A166 |
| 13-0003 | R | 13-0003R | 4qA161S | 12.000000 | 4A161S/4B163, 10A166/10A166 |
| 13-0003 | L | 13-0003L | 4qA161S | 8.000000 | 4A161S/4B163, 10A166/10A166 |
| 13-0004 | R | 13-0004R | 4qA161S | 4.000000 | 4A161S(4RU)/4A166H(27RU), 10A166/10A166 |
| 13-0004 | L | 13-0004L | 4qA161S | 8.000000 | 4A161S(4RU)/4A166H(27RU), 10A166/10A166 |
| 13-0005 | R | 13-0005R | 4qA161S | 8.000000 | 4A161S(9RU)/4B163(23RU), 10A166/10A166 |
| 13-0005 | L | 13-0005L | 4qA161S | 16.000000 | 4A161S(9RU)/4B163(23RU), 10A166/10A166 |
| 13-0006 | L | 13-0006L | 4qA161L | 8.000000 | 4A161/4A161L, 10A166/10A166 |
| 13-0007 | R | 13-0007R | 4qA161L | 42.857140 | 4A161L(2RU)/4B163(27RU), 10A164/10A166 |
| 13-0007 | L | 13-0007L | 4qA161L | 38.095240 | 4A161L(2RU)/4B163(27RU), 10A164/10A166 |
| 13-0008 | R | 13-0008R | 4qA161S | 4.000000 | 4A161S/4A161S, 10A166/10A166 |
| 13-0008 | L | 13-0008L | 4qA161S | 8.333333 | 4A161S/4A161S, 10A166/10A166 |
| 13-0009 | R | 13-0009R | 4qA161S | 4.166667 | 4A161S(5RU)/4A161L(22RU), 10A166/10A166 |
| 13-0009 | L | 13-0009L | 4qA161S | 4.000000 | 4A161S(5RU)/4A161L(22RU), 10A166/10A166 |
| 13-0010 | R | 13-0010R | 4qA161S | 4.000000 | 4A161S(8RU)/4A161S(21RU), 10A164/10A166 |
| 13-0010 | L | 13-0010L | 4qA161S | 8.000000 | 4A161S(8RU)/4A161S(21RU), 10A164/10A166 |
| 13-0011 | R | 13-0011R | 4qA161S | 12.000000 | 4A161S(6RU)/4B168(24RU), 10A166/10A166 |
| 13-0011 | L | 13-0011L | 4qA161S | 24.000000 | 4A161S(6RU)/4B168(24RU), 10A166/10A166 |
| 32-0020 | R | 32-0020R | 4qA161L | 9.523810 | 4A161L(6RU?)/4B168, 10A166/10A166 |
| 32-0020 | L | 32-0020L | 4qA161L | 4.761905 | 4A161L(6RU?)/4B168, 10A166/10A166 |
| 32-0021 | R | 32-0021R | 4qA161S | 8.000000 | 4A161S(3RU)/4B168(117RU), 10A166/10A166 |
| 32-0021 | L | 32-0021L | 4qA161S | 8.000000 | 4A161S(3RU)/4B168(117RU), 10A166/10A166 |
| 32-0022 | R | 32-0022R | 4qA161S | 4.000000 | 4A161S(7RU)/4A168(46RU), 10A166/10A166 |
| 32-0022 | L | 32-0022L | 4qA161S | 8.000000 | 4A161S(7RU)/4A168(46RU), 10A166/10A166 |
| 32-0023 | R | 32-0023R | 4qA161S | 8.000000 | 4A161S(9RU)/4B163(25RU), 10A166/10A166 |
| 32-0023 | L | 32-0023L | 4qA161S | 12.000000 | 4A161S(9RU)/4B163(25RU), 10A166/10A166 |
| 32-0024 | R | 32-0024R | 4qA161S | 8.000000 | 4A161S(9RU)/4A161S(36RU), 10A164/10A166 |
| 32-0024 | L | 32-0024L | 4qA161S | 12.000000 | 4A161S(9RU)/4A161S(36RU), 10A164/10A166 |
| 32-0025 | R | 32-0025R | 4qA161S | 12.000000 | 4A168S(4RU)/4B168(42RU), 10A164/10A166 |
| 32-0025 | L | 32-0025L | 4qA161S | 8.000000 | 4A168S(4RU)/4B168(42RU), 10A164/10A166 |
| 32-0026 | R | 32-0026R | 4qA161L | 4.761905 | 4A161L(6RU)/4B168(16RU), 10A166/10A166 |
| 32-0026 | L | 32-0026L | 4qA161L | 4.761905 | 4A161L(6RU)/4B168(16RU), 10A166/10A166 |
| 32-0027 | R | 32-0027R | 4qA161S | 16.000000 | 4A161S/4A166, 10A164/10A166 |
| 32-0027 | L | 32-0027L | 4qA161S | 20.000000 | 4A161S/4A166, 10A164/10A166 |
| 32-0028 | R | 32-0028R | 4qA161S | 5.125000 | 4A161S(5RU)/4A166H(10RU), 10A166/10A166 |
| 32-0028 | L | 32-0028L | 4qA161S | 4.000000 | 4A161S(5RU)/4A166H(10RU), 10A166/10A166 |
| 32-0029 | R | 32-0029R | 4qA161S | 0.000000 | 4A161S(5RU)/4A166H(40RU), 10A164/10A166 |
| 32-0029 | L | 32-0029L | 4qA161S | 4.000000 | 4A161S(5RU)/4A166H(40RU), 10A164/10A166 |
| 32-0030 | R | 32-0030R | 4qA161S | 0.000000 | 4A161S(4RU)/4B163(33RU), 10A166/10A166 |
| 32-0030 | L | 32-0030L | 4qA161S | 4.000000 | 4A161S(4RU)/4B163(33RU), 10A166/10A166 |
11.2 4qA161S and 4qA161L allele
Mean values of 4qA161S and 4qA161L allele:
bss %>%
group_by(`use BSSL`) %>%
summarise(mean=mean(`BSS Value`), min=min(`BSS Value`), max=max(`BSS Value`))
#> # A tibble: 2 × 4
#> `use BSSL` mean min max
#> <lgl> <dbl> <dbl> <dbl>
#> 1 FALSE 6.96 0 24
#> 2 TRUE 16.2 4.76 42.9
bss %>% dplyr::filter(!Subject=="01-0022") %>%
dplyr::mutate(allele=if_else(`use BSSL`, "4qA161L", "4qA161S")) %>%
dplyr::mutate(allele=factor(allele, levels=c("4qA161S", "4qA161L"))) %>%
ggplot(aes(x=allele, y=`BSS Value`)) +
geom_boxplot(width=0.5, outlier.shape=NA) +
geom_jitter(width=0.2, alpha=0.3, size=1, color="grey50") +
theme_minimal() +
labs(x="", y="% methylation") +
theme(axis.text.x = element_text(angle = 25, vjust = 1, hjust=1))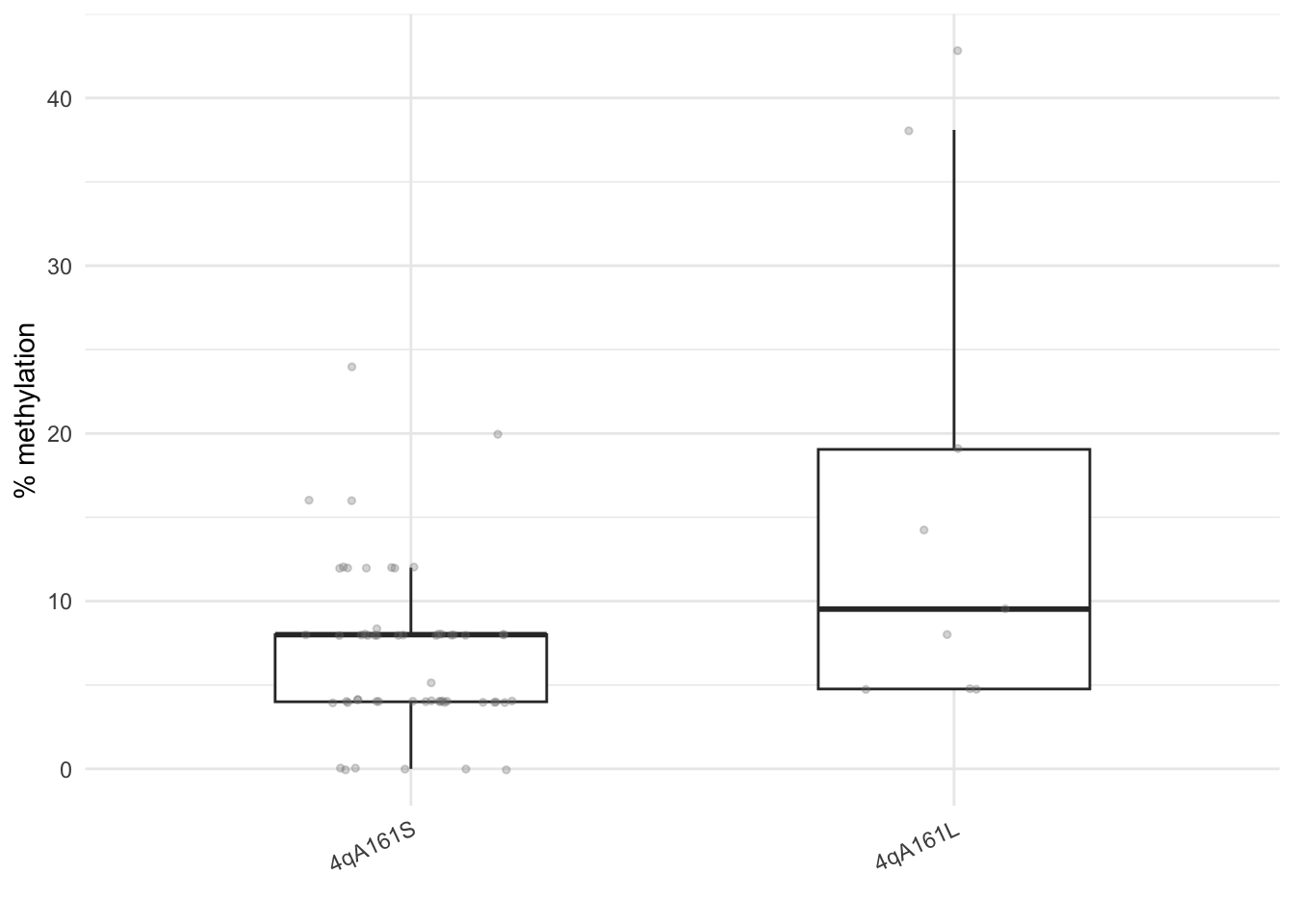
Figure 11.1: The CpG methylation percentage of subjects with 4qA161S and 4qA161L allele.
11.3 Left and right TA biopsies correlation
bss_cor <- bss %>% dplyr::select(Subject, location, `BSS Value`) %>%
spread(key=location, value=`BSS Value`) %>%
dplyr::select(-Subject) %>%
corrr::correlate() %>% dplyr::filter(!is.na(L)) %>% pull(L)
bss %>% dplyr::select(Subject, location, `BSS Value`) %>%
spread(key=location, value=`BSS Value`) %>%
ggplot(aes(x=L, y=R)) +
geom_point() +
geom_smooth(method="lm", se=FALSE, linewidth=0.7, color="grey75", alpha=0.3) +
annotate("text", x=Inf, y=-Inf, color="gray25",
size=3, hjust=1, vjust=-2,
label=paste0("Pearson=", format(bss_cor, digit=2))) +
theme_minimal() +
labs(x="L (% methylation)", y="R (% methylation)")
#> Warning: Removed 1 rows containing non-finite values
#> (`stat_smooth()`).
#> Warning: Removed 1 rows containing missing values
#> (`geom_point()`).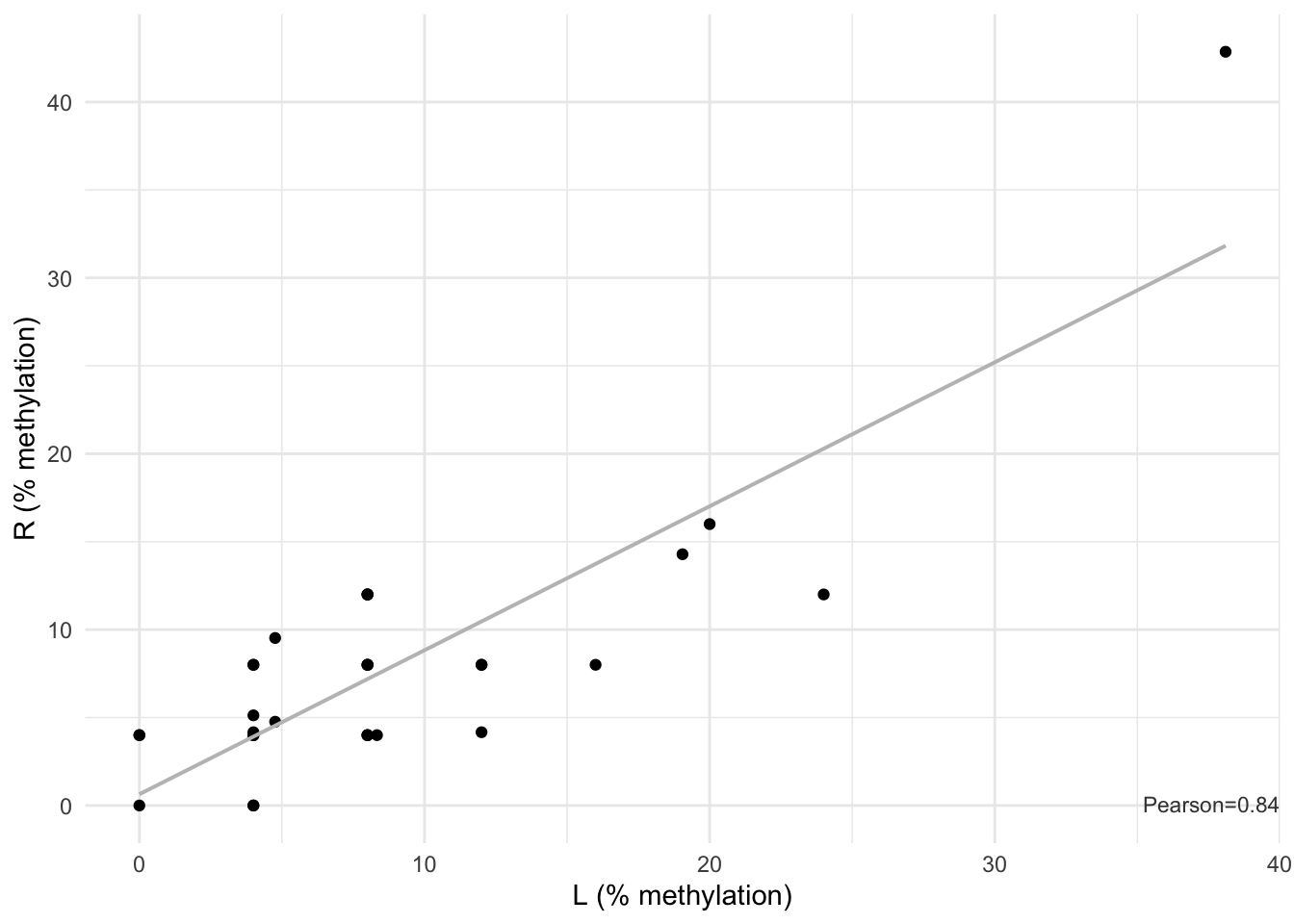
Figure 11.2: Methylation correlation between left and right TA biopsies: Pearson correlation = 0.84.
#> Warning: Removed 1 rows containing non-finite values
#> (`stat_smooth()`).
#> Warning: Removed 1 rows containing missing values
#> (`geom_point()`).11.4 Methylation and STIR status
bss %>% dplyr::select(-Subject) %>%
left_join(comprehensive_df, by="sample_id") %>%
ggplot(aes(x=STIR_status, y=`BSS Value`)) +
geom_boxplot(width=0.5, outlier.shape=NA) +
geom_jitter(width=0.3, alpha=0.3, size=1, color="grey50") +
facet_wrap(~allele) +
theme_bw() +
labs(x="", y="% methylation")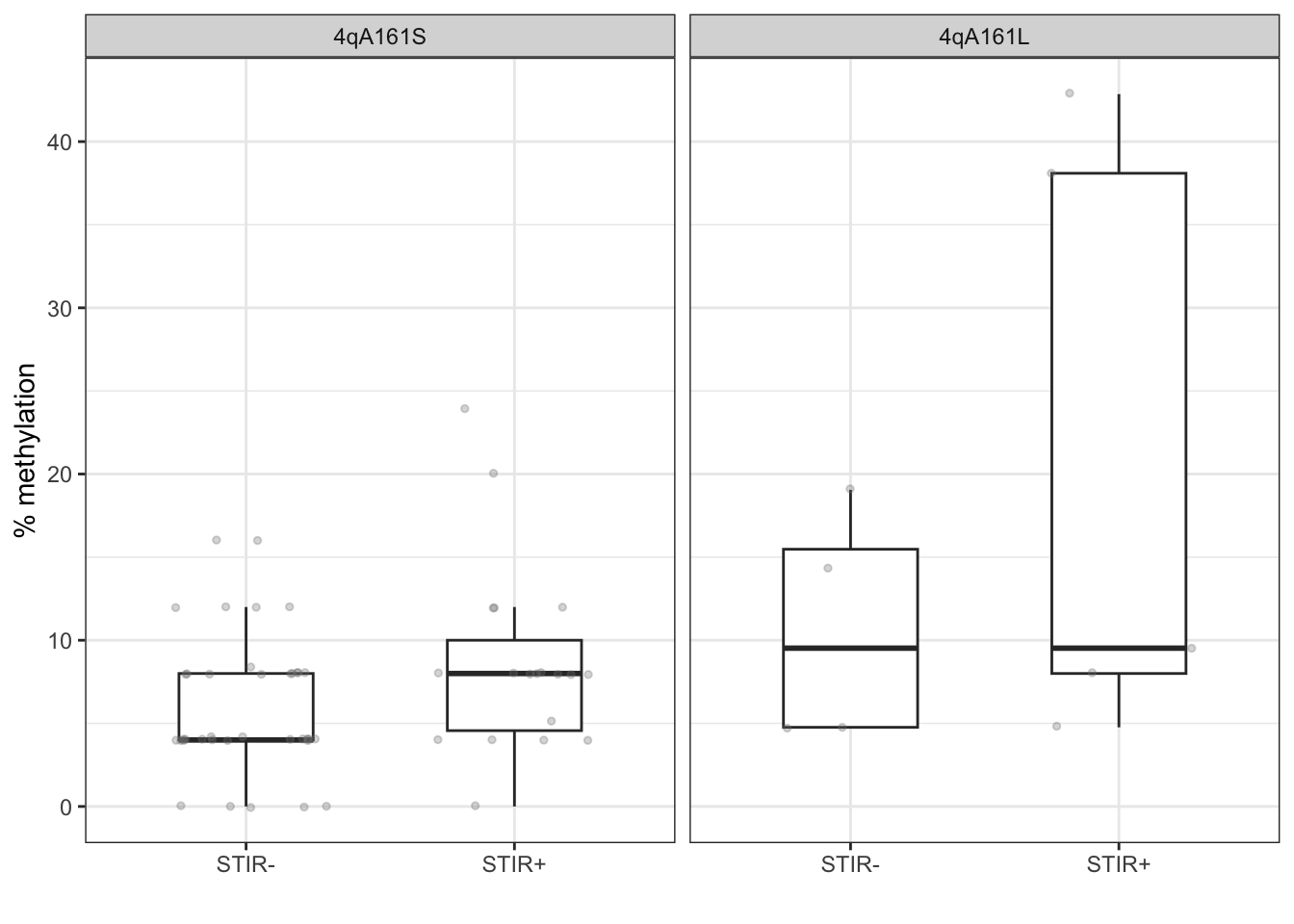
Figure 11.3: The methylation levels are not associated with STIR status.
11.5 Methylation and other variables
In the figure below we demonstrated that there is no association between CpG methylation and other variables—whole muscle fat, CSS, and muscle strength—and mRNA levels of disease signatures.
cor_161S <- bss %>% dplyr::select(-Subject) %>%
dplyr::filter(`allele`=="4qA161S") %>%
left_join(comprehensive_df, by="sample_id") %>%
dplyr::select(`BSS Value`, `DUX4-M6`, `Foot Dorsiflexors`,
`CSS`, `Fat_Infilt_Percent`, `Cumulative Score`) %>%
corrr::correlate() %>%
select(term, `BSS Value`)
cor_161L <- bss %>% dplyr::select(-Subject) %>%
dplyr::filter(`allele`=="4qA161L") %>%
left_join(comprehensive_df, by="sample_id") %>%
dplyr::select(`BSS Value`, `DUX4-M6`, `Foot Dorsiflexors`,
`CSS`, `Fat_Infilt_Percent`, `Cumulative Score`) %>%
corrr::correlate() %>%
select(term, `BSS Value`)
cor_161S %>%
full_join(cor_161L, by="term", suffix=c("4qA161S", "4qA161L")) %>%
drop_na() %>%
rename(`4qA161S`=`BSS Value4qA161S`, `4qA161L`=`BSS Value4qA161L`) %>%
gather(key=allele, value=cor, -term) %>%
ggplot(aes(x=allele, y=term)) +
geom_tile(aes(fill=cor), colour = "grey50") +
geom_text(aes(label=format(cor, digit=1)), size=3) +
scale_fill_steps2(low="grey75", mid="grey95", high="grey75",
breaks=seq(-1, 1, length.out=10)) +
theme_minimal() +
labs(x="", y="", title="Pearson correlation") +
theme(legend.position = "none",
panel.grid.major = element_blank(),
panel.grid.minor = element_blank())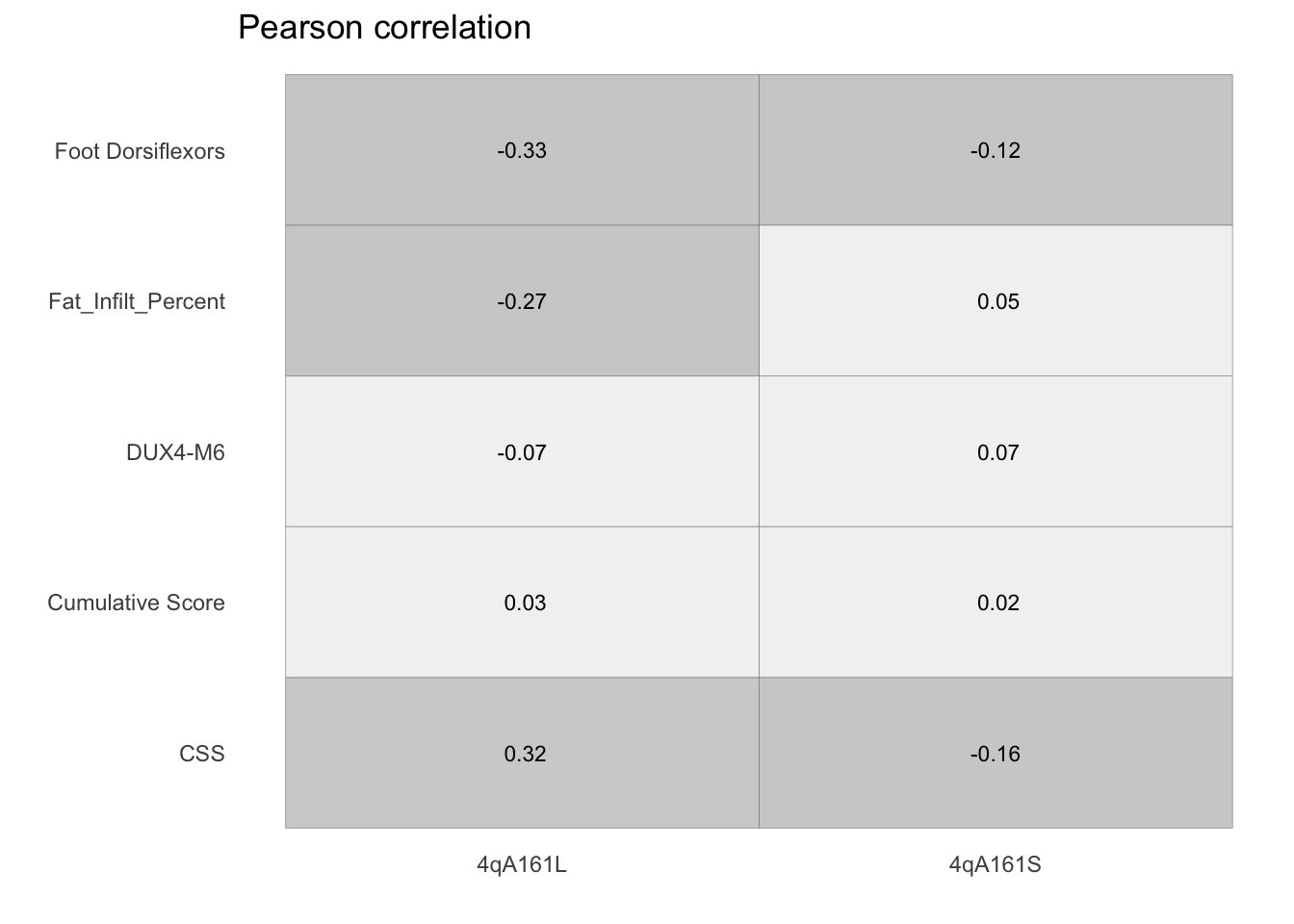
Figure 11.4: Pearson correlation between methylation levels and other variables including mRNA levels of disease signatures, whole muscle fat, CSS, and muscle strengh.